基於sklearn的整合學習實戰
整合學習投票法與bagging
投票法
sklearn提供了VotingRegressor和VotingClassifier兩個投票方法。使用模型需要提供一個模型的列表,列表中每個模型採用tuple的結構表示,第一個元素代表名稱,第二個元素代表模型,需要保證每個模型擁有唯一的名稱。看下面的例子:
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import VotingClassifier
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
models = [('lr',LogisticRegression()),('svm',SVC())]
ensemble = VotingClassifier(estimators=models) # 硬投票
models = [('lr',LogisticRegression()),('svm',make_pipeline(StandardScaler(),SVC()))]
ensemble = VotingClassifier(estimators=models,voting='soft') # 軟投票
我們可以通過一個例子來判斷整合對模型的提升效果。
首先我們建立一個1000個樣本,20個特徵的亂資料集合:
from sklearn.datasets import make_classification
def get_dataset():
X, y = make_classification(n_samples = 1000, # 樣本數目為1000
n_features = 20, # 樣本特徵總數為20
n_informative = 15, # 含有資訊量的特徵為15
n_redundant = 5, # 冗餘特徵為5
random_state = 2)
return X, y
補充一下函數make_classification的引數:
- n_samples:樣本數量,預設100
- n_features:特徵總數,預設20
- n_imformative:資訊特徵的數量
- n_redundant:冗餘特徵的數量,是資訊特徵的隨機線性組合生成的
- n_repeated:從資訊特徵和冗餘特徵中隨機抽取的重複特徵的數量
- n_classes:分類數目
- n_clusters_per_class:每個類的叢集數
- random_state:隨機種子
使用KNN模型來作為基模型:
def get_voting():
models = list()
models.append(('knn1', KNeighborsClassifier(n_neighbors=1)))
models.append(('knn3', KNeighborsClassifier(n_neighbors=3)))
models.append(('knn5', KNeighborsClassifier(n_neighbors=5)))
models.append(('knn7', KNeighborsClassifier(n_neighbors=7)))
models.append(('knn9', KNeighborsClassifier(n_neighbors=9)))
ensemble = VotingClassifier(estimators=models, voting='hard')
return ensemble
為了顯示每次模型的提升,加入下面的函數:
def get_models():
models = dict()
models['knn1'] = KNeighborsClassifier(n_neighbors=1)
models['knn3'] = KNeighborsClassifier(n_neighbors=3)
models['knn5'] = KNeighborsClassifier(n_neighbors=5)
models['knn7'] = KNeighborsClassifier(n_neighbors=7)
models['knn9'] = KNeighborsClassifier(n_neighbors=9)
models['hard_voting'] = get_voting()
return models
接下來定義下面的函數來以分層10倍交叉驗證3次重複的分數列表的形式返回:
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
def evaluate_model(model, X, y):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state = 1)
# 多次分層隨機打亂的K折交叉驗證
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1,
error_score='raise')
return scores
接著來總體呼叫一下:
from sklearn.neighbors import KNeighborsClassifier
import matplotlib.pyplot as plt
X, y = get_dataset()
models = get_models()
results, names = list(), list()
for name, model in models.items():
score = evaluate_model(model,X, y)
results.append(score)
names.append(name)
print("%s %.3f (%.3f)" % (name, score.mean(), score.std()))
plt.boxplot(results, labels = names, showmeans = True)
plt.show()
knn1 0.873 (0.030)
knn3 0.889 (0.038)
knn5 0.895 (0.031)
knn7 0.899 (0.035)
knn9 0.900 (0.033)
hard_voting 0.902 (0.034)
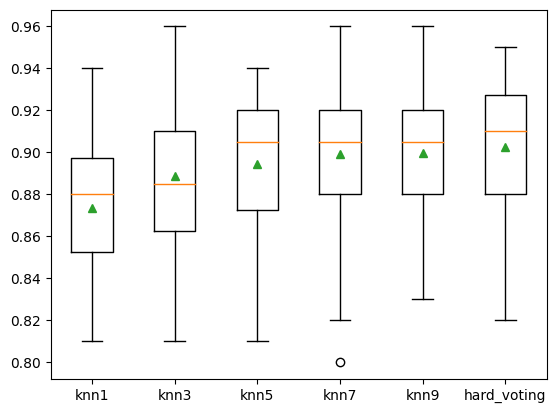
可以看到結果不斷在提升。
bagging
同樣,我們生成資料集後採用簡單的例子來介紹bagging對應函數的用法:
from numpy import mean
from numpy import std
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.ensemble import BaggingClassifier
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=5)
model = BaggingClassifier()
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Accuracy: 0.861 (0.042)
Boosting
關於這方面的理論知識的介紹可以看我這篇部落格。
這邊繼續關注這方面的程式碼怎麼使用。
Adaboost
先匯入各種包:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use("ggplot")
%matplotlib inline
import seaborn as sns
# 載入訓練資料:
wine = pd.read_csv("https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data",header=None)
wine.columns = ['Class label', 'Alcohol', 'Malic acid', 'Ash', 'Alcalinity of ash','Magnesium', 'Total phenols','Flavanoids', 'Nonflavanoid phenols',
'Proanthocyanins','Color intensity', 'Hue','OD280/OD315 of diluted wines','Proline']
# 資料檢視:
print("Class labels",np.unique(wine["Class label"]))
wine.head()
Class labels [1 2 3]
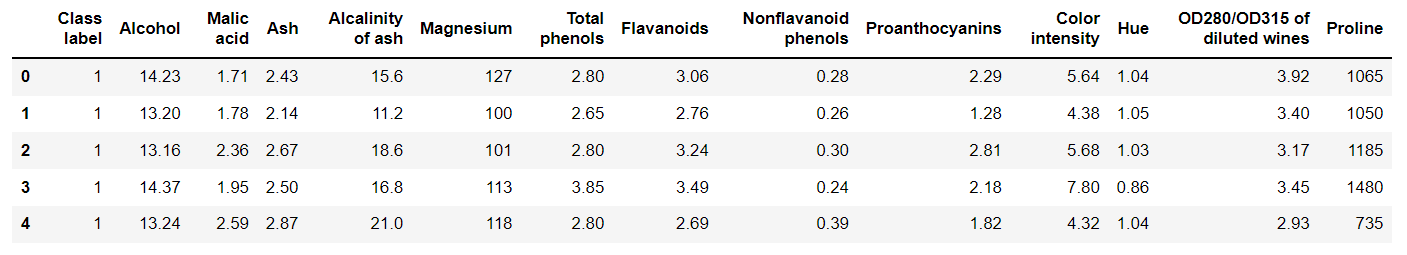
那麼接下來就需要對資料進行預處理:
# 資料預處理
# 僅僅考慮2,3類葡萄酒,去除1類
wine = wine[wine['Class label'] != 1]
y = wine['Class label'].values
X = wine[['Alcohol','OD280/OD315 of diluted wines']].values
# 將分類標籤變成二進位制編碼:
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y = le.fit_transform(y)
# 按8:2分割訓練集和測試集
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.2,random_state=1,stratify=y) # stratify引數代表了按照y的類別等比例抽樣
這裡補充一下LabelEncoder的用法,官方給出的最簡潔的解釋為:
Encode labels with value between 0 and n_classes-1.
也就是可以將原來的標籤約束到[0,n_classes-1]之間,1個數位代表一個類別。其具體的方法為:
-
fit、transform:
le = LabelEncoder() le = le.fit(['A','B','C']) # 使用le去擬合所有標籤 data = le.transform(data) # 將原來的標籤轉換為編碼 -
fit_transform:
le = LabelEncoder() data = le.fit_transform(data) -
inverse_transform:根據編碼反向推出原來類別標籤
le.inverse_transform([0,1,2]) # 輸出['A','B','C']
繼續AdaBoost。
我們接下來對比一下單一決策樹和整合之間的效果差異。
# 使用單一決策樹建模
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(criterion='entropy',random_state=1,max_depth=1)
from sklearn.metrics import accuracy_score
tree = tree.fit(X_train,y_train)
y_train_pred = tree.predict(X_train)
y_test_pred = tree.predict(X_test)
tree_train = accuracy_score(y_train,y_train_pred)
tree_test = accuracy_score(y_test,y_test_pred)
print('Decision tree train/test accuracies %.3f/%.3f' % (tree_train,tree_test))
Decision tree train/test accuracies 0.916/0.875
下面對AdaBoost進行建模:
from sklearn.ensemble import AdaBoostClassifier
ada = AdaBoostClassifier(base_estimator=tree, # 基分類器
n_estimators=500, # 最大迭代次數
learning_rate=0.1,
random_state= 1)
ada.fit(X_train, y_train)
y_train_pred = ada.predict(X_train)
y_test_pred = ada.predict(X_test)
ada_train = accuracy_score(y_train,y_train_pred)
ada_test = accuracy_score(y_test,y_test_pred)
print('Adaboost train/test accuracies %.3f/%.3f' % (ada_train,ada_test))
Adaboost train/test accuracies 1.000/0.917
可以看見分類精度提升了不少,下面我們可以觀察他們的決策邊界有什麼不同:
x_min = X_train[:, 0].min() - 1
x_max = X_train[:, 0].max() + 1
y_min = X_train[:, 1].min() - 1
y_max = X_train[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.1),np.arange(y_min, y_max, 0.1))
f, axarr = plt.subplots(nrows=1, ncols=2,sharex='col',sharey='row',figsize=(12, 6))
for idx, clf, tt in zip([0, 1],[tree, ada],['Decision tree', 'Adaboost']):
clf.fit(X_train, y_train)
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
axarr[idx].contourf(xx, yy, Z, alpha=0.3)
axarr[idx].scatter(X_train[y_train==0, 0],X_train[y_train==0, 1],c='blue', marker='^')
axarr[idx].scatter(X_train[y_train==1, 0],X_train[y_train==1, 1],c='red', marker='o')
axarr[idx].set_title(tt)
axarr[0].set_ylabel('Alcohol', fontsize=12)
plt.tight_layout()
plt.text(0, -0.2,s='OD280/OD315 of diluted wines',ha='center',va='center',fontsize=12,transform=axarr[1].transAxes)
plt.show()
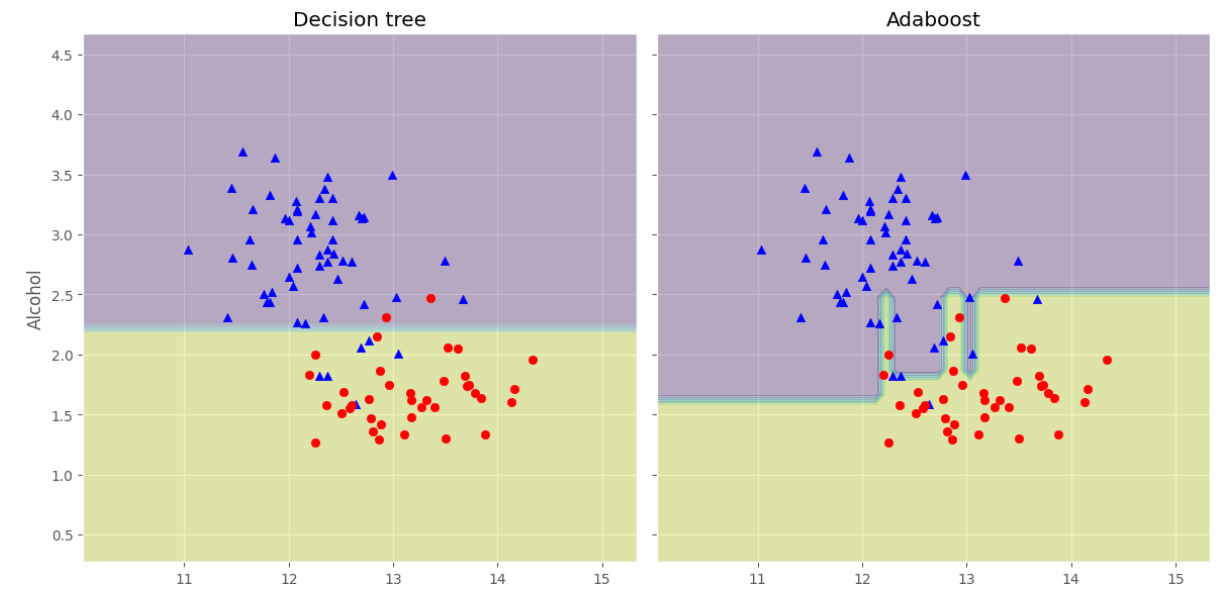
可以看淡AdaBoost的決策邊界比單個決策樹的決策邊界複雜很多。
梯度提升GBDT
這裡的理論解釋用我對西瓜書的學習筆記吧:
GBDT是一種迭代的決策樹演演算法,由多個決策樹組成,所有樹的結論累加起來作為最終答案。接下來從這兩個方面進行介紹:Regression Decision Tree(迴歸樹)、Gradient Boosting(梯度提升)、Shrinkage
DT
先補充一下決策樹的類別,包含兩種:
- 分類決策樹:用於預測分類標籤值,例如天氣的型別、使用者的性別等
- 迴歸決策樹:用來預測連續實數值,例如天氣溫度、使用者年齡
這兩者的重要區別在於迴歸決策樹的輸出結果可以相加,分類決策樹的輸出結果不可以相加,例如迴歸決策樹分別輸出10歲、5歲、2歲,那他們相加得到17歲,可以作為結果使用;但是分類決策樹分別輸出男、男、女,這樣的結果是無法進行相加處理的。因此GBDT中的樹都是迴歸樹,GBDT的核心在於累加所有樹的結果作為最終結果。
迴歸樹,救贖用樹模型來做迴歸問題,其每一片葉子都輸出一個預測值,而這個預測值一般是該片葉子所含訓練集元素輸出的均值,即$c_m=ave(y_i\mid x_i \in leaf_m)$
一般來說常見的迴歸決策樹為CART,因此下面先介紹CART如何應用於迴歸問題。
在迴歸問題中,CART主要使用均方誤差(mse)或者平均絕對誤差(mae)來作為選擇特徵以及選擇劃分點時的依據。因此用於迴歸問題時,目標就是要構建出一個函數$f(x)$能夠有效地擬合資料集$D$(樣本數為n)中的元素,使得所選取的度量指標最小,例如取mse,即:
$$
min \frac{1}{n} \sum_{i=0}{n}(f(x_i)-y_i)2
$$
而對於構建好的CART迴歸樹來說,假設其存在$M$片葉子,那麼其mse的公式可以寫成:
$$
min \frac{1}{n}\sum_{m=1}^{M}\sum_{x_i \in R_m}(c_m - y_i)^2
$$
其中$R_m$代表第$m$片葉子,而$c_m$代表第$m$片葉子的預測值。
要使得最小化mse,就需要對每一片葉子的mse都進行最小化。由統計學的知識可知道,只需要使**每片葉子的預測值為該片葉子中含有的訓練元素的均值即可,即$c_m=ave(y_i\mid x_i \in leaf_m)$。
因此CART的學習方法就是遍歷每一個變數且遍歷變數中的每一個切分點,尋找能夠使得mse最小的切分變數和切分點,即最小化如下公式:
$$
min_{j,s}[min_{c1}\sum_{x_i\in R_{1{j,s}}}(y_i-c_1)^2+min_{c_2}\sum_{x_i\in R_{2{j,s}}}(y_i-c_2)^2]
$$
另外一個值得了解的想法就是為什麼CART必須是二元樹,其實是因為如果劃分結點增多,即劃分出來的區間也增多,遍歷的難度加大。而如果想要細分為多個區域,實際上只需要讓CART迴歸樹的層次更深即可,這樣遍歷難度將小很多。
Gradient Boosting
梯度提升演演算法的流程與AdaBoost有點類似,而區別在於AdaBoost的特點在於每一輪是對分類正確和錯誤的樣本分別進行修改權重,而梯度提升演演算法的每一輪學習都是為了減少上一輪的誤差,具體可以看以下演演算法描述:
Step1、給定訓練資料集T及損失函數L,這裡可以認為損失函數為mse
Step2、初始化:
$$
f_0(x)=argmin_c\sum_{i=1}{N}L(y_i,c)=argmin_c\sum_{i=1}{N}(y_i - f_o(x_i))^2
$$
上式求解出來為:
$$
f_0(x)=\sum_{i=1}^{N} \frac{y_i}{N}=c
$$
Step3、目標基分類器(迴歸樹)數目為$M$,對於$m=1,2,...M$:
(1)、對於$i=1,2,...N$,計算
$$
r_{mi}=-[\frac{\partial L(y_i,f(x_i)}{\partial f(x_i)}]{f(x)=f{m-1}(x)}
$$
(2)、對$r_{mi}$進行擬合學習得到一個迴歸樹,其葉結點區域為$R_{mj},j=1,2,...J$
(3)、對$j=1,2,...J$,計算該對應葉結點$R_{mj}$的輸出預測值:
$$
c_{mj}=argmin_c\sum_{x_i \in R_{mj}}L(y_i,f_{m-1}(x_i)+c)
$$
(4)、更新$f_m(x)=f_{m-1}(x)+\sum {j=1}^{J}c{mj}I(x \in R_{mj})$
Step4、得到迴歸樹:
$$
\hat{f}(x)=f_M(x)=\sum_{m=0}^{M} \sum_{j=1}^{J}c_{mj}I(x\in R_{mj})
$$
為什麼損失函數的負梯度能夠作為迴歸問題殘差的近似值呢?:因為對於損失函數為mse來說,其求導的結果其實就是預測值與真實值之間的差值,那麼負梯度也就是我們預測的殘差$(y_i - f(x_i))$,因此只要下一個迴歸樹對負梯度進行擬合再對多個迴歸樹進行累加,就可以更好地逼近真實值。
Shrinkage
Shrinkage是一種用來對GBDT進行優化,防止其陷入過擬合的方法,其具體思想是:減少每一次迭代對於殘差的收斂程度或者逼近程度,也就是說該思想認為迭代時每一次少逼近一些,然後迭代次數多一些的效果,比每一次多逼近一些,然後迭代次數少一些的效果要更好。那麼具體的實現就是在每個迴歸樹的累加前乘上學習率,即:
$$
f_m(x)=f_{m-1}(x)+\gamma\sum_{j=1}^{J}c_{mj}I(x\in R_{mj})
$$
建模實現
下面對GBDT的模型進行解釋以及建模實現。
引入相關庫:
from sklearn.metrics import mean_squared_error
from sklearn.datasets import make_friedman1
from sklearn.ensemble import GradientBoostingRegressor
GBDT還有一個做分類的模型是GradientBoostingClassifier。
下面整理一下模型的各個引數:
| 引數名稱 | 引數意義 |
|---|---|
| loss | {‘ls’, ‘lad’, ‘huber’, ‘quantile’}, default=’ls’:‘ls’ 指最小二乘迴歸. ‘lad’是最小絕對偏差,是僅基於輸入變數的順序資訊的高度魯棒的損失函數;‘huber’ 是兩者的結合 |
| quantile | 允許分位數迴歸(用於alpha指定分位數) |
| learning_rate | 學習率縮小了每棵樹的貢獻learning_rate。在learning_rate和n_estimators之間需要權衡。 |
| n_estimators | 要執行的提升次數,可以認為是基分類器的數目 |
| subsample | 用於擬合各個基礎學習者的樣本比例。如果小於1.0,則將導致隨機梯度增強 |
| criterion | {'friedman_mse','mse','mae'},預設='friedman_mse':「 mse」是均方誤差,「 mae」是平均絕對誤差。預設值「 friedman_mse」通常是最好的,因為在某些情況下它可以提供更好的近似值。 |
| min_samples_split | 拆分內部節點所需的最少樣本數 |
| min_samples_leaf | 在葉節點處需要的最小樣本數 |
| min_weight_fraction_leaf | 在所有葉節點處(所有輸入樣本)的權重總和中的最小加權分數。如果未提供sample_weight,則樣本的權重相等。 |
| max_depth | 各個迴歸模型的最大深度。最大深度限制了樹中節點的數量。調整此引數以獲得最佳效能;最佳值取決於輸入變數的相互作用 |
| min_impurity_decrease | 如果節點分裂會導致雜質(損失函數)的減少大於或等於該值,則該節點將被分裂 |
| min_impurity_split | 提前停止樹木生長的閾值。如果節點的雜質高於閾值,則該節點將分裂 |
| max_features | {‘auto’, ‘sqrt’, ‘log2’},int或float:尋找最佳分割時要考慮的功能數量 |
可能各個引數一開始難以理解,但是隨著使用會加深印象的。
X, y = make_friedman1(n_samples=1200, random_state=0, noise=1.0)
X_train, X_test = X[:200], X[200:]
y_train, y_test = y[:200], y[200:]
est = GradientBoostingRegressor(n_estimators=100, learning_rate=0.1,
max_depth=1, random_state=0, loss='ls').fit(X_train, y_train)
mean_squared_error(y_test, est.predict(X_test))
5.009154859960321
from sklearn.datasets import make_regression
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.model_selection import train_test_split
X, y = make_regression(random_state=0)
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=0)
reg = GradientBoostingRegressor(random_state=0)
reg.fit(X_train, y_train)
reg.score(X_test, y_test)
XGBoost
關於XGBoost的理論,其實它跟GBDT差不多,只不過在泰勒展開時考慮了二階導函數,並在庫實現中增加了很多的優化。而關於其引數的設定也相當複雜,因此這裡簡單介紹其用法即可。
安裝XGBoost
pip install xgboost
資料介面
xgboost庫所使用的資料型別為特殊的DMatrix型別,因此其讀入檔案比較特殊:
# 1.LibSVM文字格式檔案
dtrain = xgb.DMatrix('train.svm.txt')
dtest = xgb.DMatrix('test.svm.buffer')
# 2.CSV檔案(不能含類別文字變數,如果存在文字變數請做特徵處理如one-hot)
dtrain = xgb.DMatrix('train.csv?format=csv&label_column=0')
dtest = xgb.DMatrix('test.csv?format=csv&label_column=0')
# 3.NumPy陣列
data = np.random.rand(5, 10) # 5 entities, each contains 10 features
label = np.random.randint(2, size=5) # binary target
dtrain = xgb.DMatrix(data, label=label)
# 4.scipy.sparse陣列
csr = scipy.sparse.csr_matrix((dat, (row, col)))
dtrain = xgb.DMatrix(csr)
# pandas資料框dataframe
data = pandas.DataFrame(np.arange(12).reshape((4,3)), columns=['a', 'b', 'c'])
label = pandas.DataFrame(np.random.randint(2, size=4))
dtrain = xgb.DMatrix(data, label=label)
第一次讀入後可以先儲存為對應的二進位制檔案方便下次讀取:
# 1.儲存DMatrix到XGBoost二進位制檔案中
dtrain = xgb.DMatrix('train.svm.txt')
dtrain.save_binary('train.buffer')
# 2. 缺少的值可以用DMatrix建構函式中的預設值替換:
dtrain = xgb.DMatrix(data, label=label, missing=-999.0)
# 3.可以在需要時設定權重:
w = np.random.rand(5, 1)
dtrain = xgb.DMatrix(data, label=label, missing=-999.0, weight=w)
引數設定
import pandas as pd
df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data',header=None)
df_wine.columns = ['Class label', 'Alcohol','Malic acid', 'Ash','Alcalinity of ash','Magnesium', 'Total phenols',
'Flavanoids', 'Nonflavanoid phenols','Proanthocyanins','Color intensity', 'Hue','OD280/OD315 of diluted wines','Proline']
df_wine = df_wine[df_wine['Class label'] != 1] # drop 1 class
y = df_wine['Class label'].values
X = df_wine[['Alcohol','OD280/OD315 of diluted wines']].values
from sklearn.model_selection import train_test_split # 切分訓練集與測試集
from sklearn.preprocessing import LabelEncoder # 標籤化分類變數
import xgboost as xgb
le = LabelEncoder()
y = le.fit_transform(y)
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.2,
random_state=1,stratify=y)
# 構造成目標型別的資料
dtrain = xgb.DMatrix(X_train, label = y_train)
dtest = xgb.DMatrix(X_test)
# booster引數
params = {
"booster": 'gbtree', # 基分類器
"objective": "multi:softmax", # 使用softmax
"num_class": 10, # 類別數
"gamma":0.1, # 用於控制損失函數大於gamma時就剪枝
"max_depth": 12, # 構造樹的深度,越大越容易過擬合
"lambda": 2, # 用來控制正則化引數
"subsample": 0.7, # 隨機取樣70%作為訓練樣本
"colsample_bytree": 0.7, # 生成樹時進行列取樣,相當於隨機子空間
"min_child_weight":3,
'verbosity':0, # 設定成1則沒有執行資訊輸出,設定成0則有,原文中這裡的silent已經在新版本中不使用了
"eta": 0.007, # 相當於學習率
"seed": 1000, # 亂數種子
"nthread":4, # 執行緒數
"eval_metric": "auc"
}
plst = list(params.items()) # 這裡必須加上list才能夠使用後續的copy
這裡如果發現報錯為:
Parameters: { "silent" } are not used.
那是因為引數設定中新版本已經取消了silent引數,將其更改為verbosity即可。
如果在後續中發現:
'dict_items' object has no attribute 'copy'
那是因為我們沒有將items的返回變成list,像我上面那麼改即可。
訓練及預測
num_round = 10
bst = xgb.train(plst, dtrain ,num_round)
# 可以加上early_stoppint_rounds = 10來設定早停機制
# 如果要指定驗證集,就是
# evallist = [(deval,'eval'),(dtrain, 'train')]
# bst = xgb.train(plst, dtrain, num_round, evallist)
ypred = bst.predict(dtest) # 進行預測,跟sklearn的模型一致
xgb.plot_importance(bst) # 繪製特徵重要性的圖
<AxesSubplot:title={'center':'Feature importance'}, xlabel='F score', ylabel='Features'>
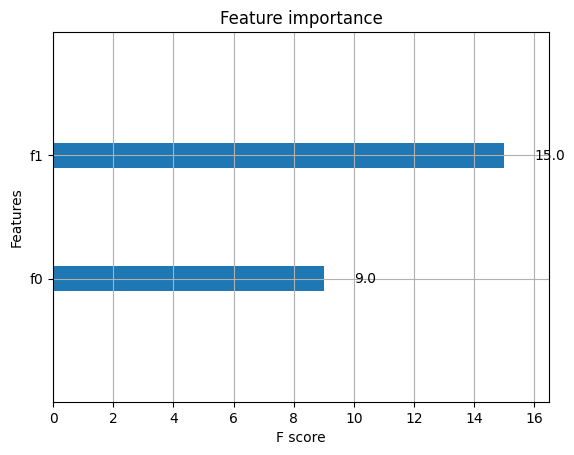
模型儲存與載入
bst.save_model("model_new_1121.model")
bst.dump_model("dump.raw.txt")
bst_new = xgb.Booster({'nthread':4}) # 先初始化引數
bst_new.load_model("model_new_1121.model")
簡單算例
分類案例
from sklearn.datasets import load_iris
import xgboost as xgb
from xgboost import plot_importance
from matplotlib import pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score # 準確率
# 載入樣本資料集
iris = load_iris()
X,y = iris.data,iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2,
random_state=1234565) # 資料集分割
# 演演算法引數
params = {
'booster': 'gbtree',
'objective': 'multi:softmax',
'num_class': 3,
'gamma': 0.1,
'max_depth': 6,
'lambda': 2,
'subsample': 0.7,
'colsample_bytree': 0.75,
'min_child_weight': 3,
'verbosity': 0,
'eta': 0.1,
'seed': 1,
'nthread': 4,
}
plst = list(params.items())
dtrain = xgb.DMatrix(X_train, y_train) # 生成資料集格式
num_rounds = 500
model = xgb.train(plst, dtrain, num_rounds) # xgboost模型訓練
# 對測試集進行預測
dtest = xgb.DMatrix(X_test)
y_pred = model.predict(dtest)
# 計算準確率
accuracy = accuracy_score(y_test,y_pred)
print("accuarcy: %.2f%%" % (accuracy*100.0))
# 顯示重要特徵
plot_importance(model)
plt.show()
accuarcy: 96.67%
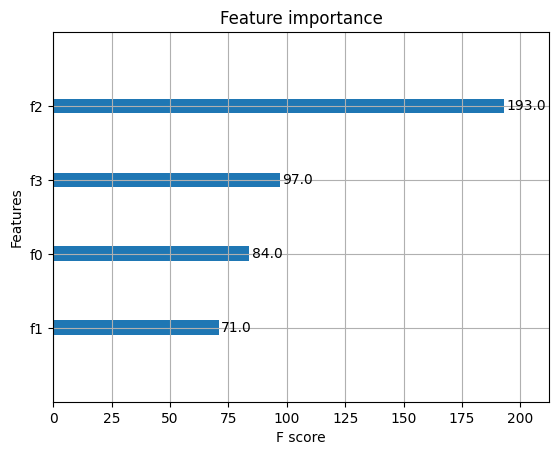
迴歸案例
import xgboost as xgb
from xgboost import plot_importance
from matplotlib import pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.datasets import load_boston
from sklearn.metrics import mean_squared_error
# 載入資料集
boston = load_boston()
X,y = boston.data,boston.target
# XGBoost訓練過程
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
params = {
'booster': 'gbtree',
'objective': 'reg:squarederror', # 設定為迴歸,採用平方誤差
'gamma': 0.1,
'max_depth': 5,
'lambda': 3,
'subsample': 0.7,
'colsample_bytree': 0.7,
'min_child_weight': 3,
'verbosity': 1,
'eta': 0.1,
'seed': 1000,
'nthread': 4,
}
dtrain = xgb.DMatrix(X_train, y_train)
num_rounds = 300
plst = list(params.items())
model = xgb.train(plst, dtrain, num_rounds)
# 對測試集進行預測
dtest = xgb.DMatrix(X_test)
ans = model.predict(dtest)
# 顯示重要特徵
plot_importance(model)
plt.show()
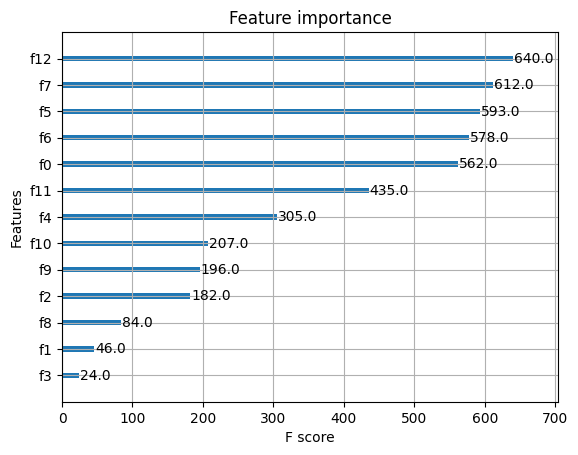
XGBoost的調參
該模型的調參一般步驟為:
- 確定學習速率和提升引數調優的初始值
- max_depth 和 min_child_weight 引數調優
- gamma引數調優
- subsample 和 colsample_bytree 引數優
- 正則化引數alpha調優
- 降低學習速率和使用更多的決策樹
可以使用網格搜尋來進行調優:
import xgboost as xgb
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import roc_auc_score
iris = load_iris()
X,y = iris.data,iris.target
col = iris.target_names
train_x, valid_x, train_y, valid_y = train_test_split(X, y, test_size=0.3, random_state=1) # 分訓練集和驗證集
parameters = {
'max_depth': [5, 10, 15, 20, 25],
'learning_rate': [0.01, 0.02, 0.05, 0.1, 0.15],
'n_estimators': [500, 1000, 2000, 3000, 5000],
'min_child_weight': [0, 2, 5, 10, 20],
'max_delta_step': [0, 0.2, 0.6, 1, 2],
'subsample': [0.6, 0.7, 0.8, 0.85, 0.95],
'colsample_bytree': [0.5, 0.6, 0.7, 0.8, 0.9],
'reg_alpha': [0, 0.25, 0.5, 0.75, 1],
'reg_lambda': [0.2, 0.4, 0.6, 0.8, 1],
'scale_pos_weight': [0.2, 0.4, 0.6, 0.8, 1]
}
xlf = xgb.XGBClassifier(max_depth=10,
learning_rate=0.01,
n_estimators=2000,
silent=True,
objective='multi:softmax',
num_class=3 ,
nthread=-1,
gamma=0,
min_child_weight=1,
max_delta_step=0,
subsample=0.85,
colsample_bytree=0.7,
colsample_bylevel=1,
reg_alpha=0,
reg_lambda=1,
scale_pos_weight=1,
seed=0,
missing=None)
# 需要先給模型一定的初始值
gs = GridSearchCV(xlf, param_grid=parameters, scoring='accuracy', cv=3)
gs.fit(train_x, train_y)
print("Best score: %0.3f" % gs.best_score_)
print("Best parameters set: %s" % gs.best_params_ )
Best score: 0.933
Best parameters set: {'max_depth': 5}
LightGBM演演算法
LightGBM也是像XGBoost一樣,是一類整合演演算法,他跟XGBoost總體來說是一樣的,演演算法本質上與Xgboost沒有出入,只是在XGBoost的基礎上進行了優化。
其調優過程也是一個很複雜的學問。這裡就附上課程調優程式碼吧:
import lightgbm as lgb
from sklearn import metrics
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
canceData=load_breast_cancer()
X=canceData.data
y=canceData.target
X_train,X_test,y_train,y_test=train_test_split(X,y,random_state=0,test_size=0.2)
### 資料轉換
print('資料轉換')
lgb_train = lgb.Dataset(X_train, y_train, free_raw_data=False)
lgb_eval = lgb.Dataset(X_test, y_test, reference=lgb_train,free_raw_data=False)
### 設定初始引數--不含交叉驗證引數
print('設定引數')
params = {
'boosting_type': 'gbdt',
'objective': 'binary',
'metric': 'auc',
'nthread':4,
'learning_rate':0.1
}
### 交叉驗證(調參)
print('交叉驗證')
max_auc = float('0')
best_params = {}
# 準確率
print("調參1:提高準確率")
for num_leaves in range(5,100,5):
for max_depth in range(3,8,1):
params['num_leaves'] = num_leaves
params['max_depth'] = max_depth
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['num_leaves'] = num_leaves
best_params['max_depth'] = max_depth
if 'num_leaves' and 'max_depth' in best_params.keys():
params['num_leaves'] = best_params['num_leaves']
params['max_depth'] = best_params['max_depth']
# 過擬合
print("調參2:降低過擬合")
for max_bin in range(5,256,10):
for min_data_in_leaf in range(1,102,10):
params['max_bin'] = max_bin
params['min_data_in_leaf'] = min_data_in_leaf
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['max_bin']= max_bin
best_params['min_data_in_leaf'] = min_data_in_leaf
if 'max_bin' and 'min_data_in_leaf' in best_params.keys():
params['min_data_in_leaf'] = best_params['min_data_in_leaf']
params['max_bin'] = best_params['max_bin']
print("調參3:降低過擬合")
for feature_fraction in [0.6,0.7,0.8,0.9,1.0]:
for bagging_fraction in [0.6,0.7,0.8,0.9,1.0]:
for bagging_freq in range(0,50,5):
params['feature_fraction'] = feature_fraction
params['bagging_fraction'] = bagging_fraction
params['bagging_freq'] = bagging_freq
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc=mean_auc
best_params['feature_fraction'] = feature_fraction
best_params['bagging_fraction'] = bagging_fraction
best_params['bagging_freq'] = bagging_freq
if 'feature_fraction' and 'bagging_fraction' and 'bagging_freq' in best_params.keys():
params['feature_fraction'] = best_params['feature_fraction']
params['bagging_fraction'] = best_params['bagging_fraction']
params['bagging_freq'] = best_params['bagging_freq']
print("調參4:降低過擬合")
for lambda_l1 in [1e-5,1e-3,1e-1,0.0,0.1,0.3,0.5,0.7,0.9,1.0]:
for lambda_l2 in [1e-5,1e-3,1e-1,0.0,0.1,0.4,0.6,0.7,0.9,1.0]:
params['lambda_l1'] = lambda_l1
params['lambda_l2'] = lambda_l2
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc=mean_auc
best_params['lambda_l1'] = lambda_l1
best_params['lambda_l2'] = lambda_l2
if 'lambda_l1' and 'lambda_l2' in best_params.keys():
params['lambda_l1'] = best_params['lambda_l1']
params['lambda_l2'] = best_params['lambda_l2']
print("調參5:降低過擬合2")
for min_split_gain in [0.0,0.1,0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,1.0]:
params['min_split_gain'] = min_split_gain
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc=mean_auc
best_params['min_split_gain'] = min_split_gain
if 'min_split_gain' in best_params.keys():
params['min_split_gain'] = best_params['min_split_gain']
print(best_params)
{'bagging_fraction': 0.7,
'bagging_freq': 30,
'feature_fraction': 0.8,
'lambda_l1': 0.1,
'lambda_l2': 0.0,
'max_bin': 255,
'max_depth': 4,
'min_data_in_leaf': 81,
'min_split_gain': 0.1,
'num_leaves': 10}
Blending與Stacking
Stacking,這個整合方法在比賽中被稱為「懶人」演演算法,因為它不需要花費過多時間的調參就可以得到一個效果不錯的演演算法。
Stacking整合演演算法可以理解為一個兩層的整合,第一層含有多個基礎分類器,把預測的結果(元特徵)提供給第二層, 而第二層的分類器通常是邏輯迴歸,他把一層分類器的結果當做特徵做擬合輸出預測結果。
而Blending就是簡化版的Stacking,因此我們先對前者進行介紹。
Blending整合學習演演算法
演演算法流程
Blending的演演算法流程為:
- 將資料集劃分為訓練集與測試集,訓練集再劃分為訓練集與驗證集
- 建立第一層的多個模型(可同質也可異質),然後對訓練集進行學習
- 第一層的模型訓練完成後對驗證集和測試集做出預測,假設K個模型,那麼就得到$A_1,...,A_K$和$B_1,...,B_K$,其中每個代表一個基分類器對驗證集或測試集的所有結果輸出。
- 建立第二層的分類器,其將$A_1,...,A_K$作為訓練資料集,那麼樣本數目就是驗證集的樣本數目,特徵數目就是K,將真實的驗證集標籤作為標籤,從而來訓練該分類器
- 對測試集的預測則是將$B_1,...,B_K$作為特徵,用第二層的分類器進行預測。
具體實現
具體的實現如下:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use("ggplot")
%matplotlib inline
import seaborn as sns
# 建立資料
from sklearn import datasets
from sklearn.datasets import make_blobs
from sklearn.model_selection import train_test_split
data, target = make_blobs(n_samples=10000, centers=2, random_state=1, cluster_std=1.0 )
## 建立訓練集和測試集
X_train1,X_test,y_train1,y_test = train_test_split(data, target, test_size=0.2, random_state=1)
## 建立訓練集和驗證集
X_train,X_val,y_train,y_val = train_test_split(X_train1, y_train1, test_size=0.3, random_state=1)
print("The shape of training X:",X_train.shape)
print("The shape of training y:",y_train.shape)
print("The shape of test X:",X_test.shape)
print("The shape of test y:",y_test.shape)
print("The shape of validation X:",X_val.shape)
print("The shape of validation y:",y_val.shape)
The shape of training X: (5600, 2)
The shape of training y: (5600,)
The shape of test X: (2000, 2)
The shape of test y: (2000,)
The shape of validation X: (2400, 2)
The shape of validation y: (2400,)
# 設定第一層分類器
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
clfs = [SVC(probability = True),RandomForestClassifier(n_estimators=5, n_jobs=-1, criterion='gini'),KNeighborsClassifier()]
# 設定第二層分類器
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
# 輸出第一層的驗證集結果與測試集結果
val_features = np.zeros((X_val.shape[0],len(clfs))) # 初始化驗證集結果
test_features = np.zeros((X_test.shape[0],len(clfs))) # 初始化測試集結果
for i,clf in enumerate(clfs):
clf.fit(X_train,y_train)
# porba函數得到的是對於每個類別的預測分數,取出第一列代表每個樣本為第一類的概率
val_feature = clf.predict_proba(X_val)[:, 1]
test_feature = clf.predict_proba(X_test)[:,1]
val_features[:,i] = val_feature
test_features[:,i] = test_feature
# 將第一層的驗證集的結果輸入第二層訓練第二層分類器
lr.fit(val_features,y_val)
# 輸出預測的結果
from sklearn.model_selection import cross_val_score
cross_val_score(lr,test_features,y_test,cv=5)
array([1., 1., 1., 1., 1.])
可以看到交叉驗證的結果很好。
對於小作業我總有些疑問,那就是這個iris資料的特徵為4,然後預測類別數為3,那麼首先是特徵為4超過3維度,應該怎麼決策邊界,難道捨棄一些維度嗎?其次是類別數為3,那麼在計算的時候取的是[;1]也就是類別為1的概率,那麼只取這個作為下一層的特徵是否足夠,因為類別為0和類別為2的概率完全捨棄的話不行吧。
Stacking
演演算法流程
下面這張圖可以很好的理解流程:
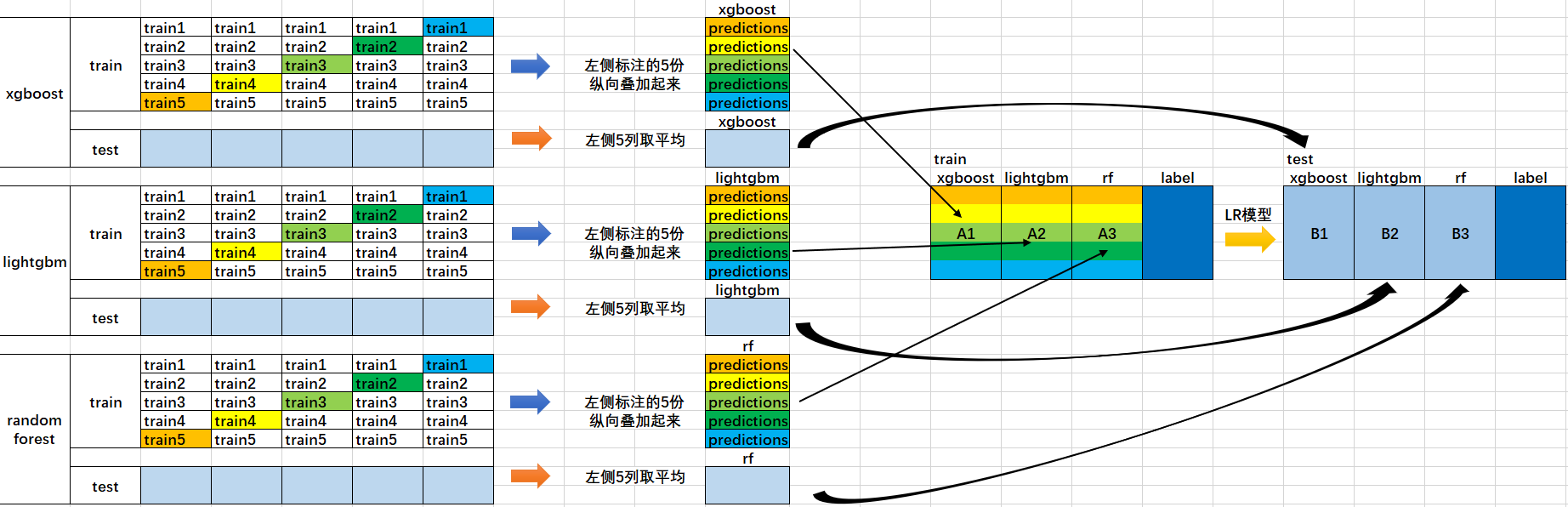
- 首先將所有資料劃分測試集和訓練集,假設訓練集總共有10000行,測試集總共2500行,對第一層的分類器進行5折交叉驗證,那麼驗證集就劃分為2000行,訓練集為8000行。
- 每次驗證就相當於使用8000條訓練資料去訓練模型然後用2000條驗證資料去驗證,並且每一次都將訓練出來的模型對測試集的2500條資料進行預測,那麼經過5次交叉驗證,對於每個基分類器可以得到中間的$5\times 2000$的五份驗證集資料,以及$5\times 2500$的五份測試集的預測結果
- 接下來將驗證集拼成10000行長的矩陣,記為$A_1$(對於第1個基分類器),而對於$5\times 2500$行的測試集的預測結果進行加權平均,得到2500行1列的矩陣,記為$B_1$
- 那麼假設這裡是3個基分類器,因此有$A_1,A_2,A_3,B_1,B_2,B_3$六個矩陣,接下來將$A_1,A_2,A_3$矩陣拼成10000行3列的矩陣作為訓練資料集,而驗證集的真實標籤作為訓練資料的標籤;將$B_1,B_2,B_3$拼成2500行3列的矩陣作為測試資料集合。
- 那麼對下層的學習器進行訓練
具體程式碼
from sklearn import datasets
iris = datasets.load_iris()
X, y = iris.data[:, 1:3], iris.target # 只取出兩個特徵
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from mlxtend.classifier import StackingCVClassifier
RANDOM_SEED = 42
clf1 = KNeighborsClassifier(n_neighbors = 1)
clf2 = RandomForestClassifier(random_state = RANDOM_SEED)
clf3 = GaussianNB()
lr = LogisticRegression()
sclf = StackingCVClassifier(classifiers = [clf1, clf2, clf3], # 第一層的分類器
meta_classifier = lr, # 第二層的分類器
random_state = RANDOM_SEED)
print("3折交叉驗證:\n")
for clf, label in zip([clf1, clf2, clf3, sclf], ['KNN','RandomForest','Naive Bayes',
'Stack']):
scores = cross_val_score(clf, X, y, cv = 3, scoring = 'accuracy')
print("Accuracy: %0.2f (+/- %0.2f) [%s]" % (scores.mean(), scores.std(), label))
3折交叉驗證:
Accuracy: 0.91 (+/- 0.01) [KNN]
Accuracy: 0.95 (+/- 0.01) [RandomForest]
Accuracy: 0.91 (+/- 0.02) [Naive Bayes]
Accuracy: 0.93 (+/- 0.02) [Stack]
接下來嘗試將決策邊界畫出:
from mlxtend.plotting import plot_decision_regions
import matplotlib.gridspec as gridspec
import itertools
gs = gridspec.GridSpec(2,2)
# 可以理解為將子圖劃分為了2*2的區域
fig = plt.figure(figsize = (10,8))
for clf, lab, grd in zip([clf1, clf2, clf3, sclf], ['KNN',
'RandomForest',
'Naive Bayes',
'Stack'],
itertools.product([0,1],repeat=2)):
clf.fit(X, y)
ax = plt.subplot(gs[grd[0],grd[1]])
# grd依次為(0,0),(0,1),(1,0),(1,1),那麼傳入到gs中就可以得到指定的區域
fig = plot_decision_regions(X = X, y = y, clf = clf)
plt.title(lab)
plt.show()
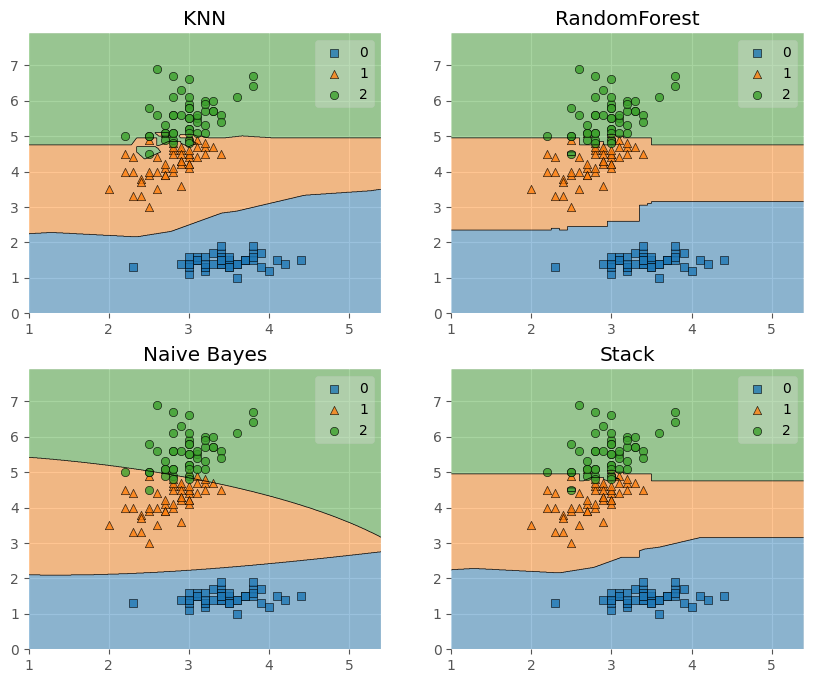
這裡補充兩個第一次見到的函數:
-
itertools.product([0,1],repeat = 2):該模組下的product函數一般是傳進入兩個集合,例如傳進入[0,1],[1,2]然後返回[(0,1),(0,2),(1,1),(1,2)],那麼這裡只傳進去一個引數然後repeat=2相當於傳進去[0,1],[0,1],產生[(0,0),(0,1),(1,0),(1,1)],如果repeat=3就是
(0, 0, 0) (0, 0, 1) (0, 1, 0) (0, 1, 1) (1, 0, 0) (1, 0, 1) (1, 1, 0) (1, 1, 1) -
gs = gridspec.GridSpec(2,2):這個函數相當於我將子圖劃分為2*2總共4個區域,那麼在下面subplot中就可以例如呼叫gs[0,1]來獲取(0,1)這個區域,下面的例子或者更好理解:
plt.figure() gs=gridspec.GridSpec(3,3)#分為3行3列 ax1=plt.subplot(gs[0,:]) ax1=plt.subplot(gs[1,:2]) ax1=plt.subplot(gs[1:,2]) ax1=plt.subplot(gs[-1,0]) ax1=plt.subplot(gs[-1,-2])
繼續回到Stacking中。
前面我們是使用第一層的分類器其輸出作為第二層的輸入,那麼如果希望使用第一層所有基分類器所產生的的類別概率值作為第二層分類器的數目,需要在StackingClassifier 中增加一個引數設定:use_probas = True。還有一個引數設定average_probas = True,那麼這些基分類器所產出的概率值將按照列被平均,否則會拼接
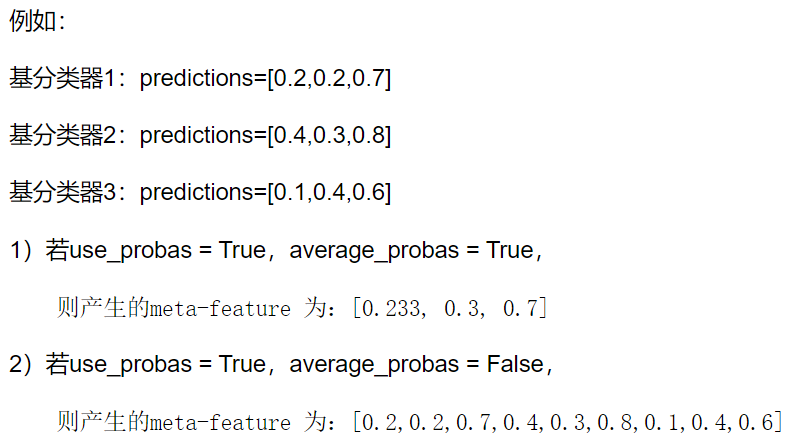
我們來進行嘗試:
clf1 = KNeighborsClassifier(n_neighbors=1)
clf2 = RandomForestClassifier(random_state=1)
clf3 = GaussianNB()
lr = LogisticRegression()
sclf = StackingCVClassifier(classifiers=[clf1, clf2, clf3],
use_probas=True, # 使用概率
meta_classifier=lr,
random_state=42)
print('3折交叉驗證:')
for clf, label in zip([clf1, clf2, clf3, sclf],
['KNN',
'Random Forest',
'Naive Bayes',
'StackingClassifier']):
scores = cross_val_score(clf, X, y, cv=3, scoring='accuracy')
print("Accuracy: %0.2f (+/- %0.2f) [%s]"
% (scores.mean(), scores.std(), label))
3折交叉驗證:
Accuracy: 0.91 (+/- 0.01) [KNN]
Accuracy: 0.95 (+/- 0.01) [Random Forest]
Accuracy: 0.91 (+/- 0.02) [Naive Bayes]
Accuracy: 0.95 (+/- 0.02) [StackingClassifier]
另外,還可以跟網格搜尋相結合:
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from mlxtend.classifier import StackingCVClassifier
clf1 = KNeighborsClassifier(n_neighbors=1)
clf2 = RandomForestClassifier(random_state=RANDOM_SEED)
clf3 = GaussianNB()
lr = LogisticRegression()
sclf = StackingCVClassifier(classifiers=[clf1, clf2, clf3],
meta_classifier=lr,
random_state=42)
params = {'kneighborsclassifier__n_neighbors': [1, 5],
'randomforestclassifier__n_estimators': [10, 50],
'meta_classifier__C': [0.1, 10.0]}#
grid = GridSearchCV(estimator=sclf, # 分類為stacking
param_grid=params, # 設定的引數
cv=5,
refit=True)
#最後一個引數代表在搜尋引數結束後,用最佳引數結果再次fit一遍全部資料集
grid.fit(X, y)
cv_keys = ('mean_test_score', 'std_test_score', 'params')
for r,_ in enumerate(grid.cv_results_['mean_test_score']):
print("%0.3f +/- %0.2f %r"
% (grid.cv_results_[cv_keys[0]][r],
grid.cv_results_[cv_keys[1]][r] / 2.0,
grid.cv_results_[cv_keys[2]][r]))
print('Best parameters: %s' % grid.best_params_)
print('Accuracy: %.2f' % grid.best_score_)
0.947 +/- 0.03 {'kneighborsclassifier__n_neighbors': 1, 'meta_classifier__C': 0.1, 'randomforestclassifier__n_estimators': 10}
0.933 +/- 0.02 {'kneighborsclassifier__n_neighbors': 1, 'meta_classifier__C': 0.1, 'randomforestclassifier__n_estimators': 50}
0.940 +/- 0.02 {'kneighborsclassifier__n_neighbors': 1, 'meta_classifier__C': 10.0, 'randomforestclassifier__n_estimators': 10}
0.940 +/- 0.02 {'kneighborsclassifier__n_neighbors': 1, 'meta_classifier__C': 10.0, 'randomforestclassifier__n_estimators': 50}
0.953 +/- 0.02 {'kneighborsclassifier__n_neighbors': 5, 'meta_classifier__C': 0.1, 'randomforestclassifier__n_estimators': 10}
0.953 +/- 0.02 {'kneighborsclassifier__n_neighbors': 5, 'meta_classifier__C': 0.1, 'randomforestclassifier__n_estimators': 50}
0.953 +/- 0.02 {'kneighborsclassifier__n_neighbors': 5, 'meta_classifier__C': 10.0, 'randomforestclassifier__n_estimators': 10}
0.953 +/- 0.02 {'kneighborsclassifier__n_neighbors': 5, 'meta_classifier__C': 10.0, 'randomforestclassifier__n_estimators': 50}
Best parameters: {'kneighborsclassifier__n_neighbors': 5, 'meta_classifier__C': 0.1, 'randomforestclassifier__n_estimators': 10}
Accuracy: 0.95
而如果希望在演演算法中多次使用某個模型,就可以在引數網格中新增一個附加的數位字尾:
params = {'kneighborsclassifier-1__n_neighbors': [1, 5],
'kneighborsclassifier-2__n_neighbors': [1, 5],
'randomforestclassifier__n_estimators': [10, 50],
'meta_classifier__C': [0.1, 10.0]}
我們還可以結合隨機子空間的思想,為Stacking第一層的不同子模型設定不同的特徵:
from sklearn.datasets import load_iris
from mlxtend.classifier import StackingCVClassifier
from mlxtend.feature_selection import ColumnSelector
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LogisticRegression
iris = load_iris()
X = iris.data
y = iris.target
pipe1 = make_pipeline(ColumnSelector(cols=(0, 2)), # 選擇第0,2列
LogisticRegression()) # 可以理解為先挑選特徵再以基分類器為邏輯迴歸
pipe2 = make_pipeline(ColumnSelector(cols=(1, 2, 3)), # 選擇第1,2,3列
LogisticRegression()) # 兩個基分類器都是邏輯迴歸
sclf = StackingCVClassifier(classifiers=[pipe1, pipe2],
# 兩個基分類器區別在於使用特徵不同
meta_classifier=LogisticRegression(),
random_state=42)
sclf.fit(X, y)

下面我們可以畫ROC曲線:
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from mlxtend.classifier import StackingCVClassifier
from sklearn.metrics import roc_curve, auc
from sklearn.model_selection import train_test_split
from sklearn import datasets
from sklearn.preprocessing import label_binarize
from sklearn.multiclass import OneVsRestClassifier
iris = datasets.load_iris()
X, y = iris.data[:, [0, 1]], iris.target # 只用了前兩個特徵
y = label_binarize(y, classes = [0,1,2])
# 因為y裡面有三個類別,分類標註為0,1,2,這裡是將y變換為一個n*3的矩陣
# 每一行為3代表類別數目為3,然後如果y是第0類就是100,第一類就是010,第二類就是001
# 關鍵在於後面的classes如何制定,如果是[0,2,1]那麼是第二類就是010,第一類是001
n_classes = y.shape[1]
RANDOM_SEED = 42
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33,
random_state=RANDOM_SEED)
clf1 = LogisticRegression()
clf2 = RandomForestClassifier(random_state=RANDOM_SEED)
clf3 = SVC(random_state=RANDOM_SEED)
lr = LogisticRegression()
sclf = StackingCVClassifier(classifiers=[clf1, clf2, clf3],
meta_classifier=lr)
classifier = OneVsRestClassifier(sclf)
# 這個物件在擬合時會對每一類學習一個分類器,用來做二分類,分別該類和其他所有類
y_score = classifier.fit(X_train, y_train).decision_function(X_test)
# decision_function是預測X_test在決策邊界的哪一邊,然後距離有多大,可以認為是評估指標
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], y_score[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), y_score.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
plt.figure()
lw = 2
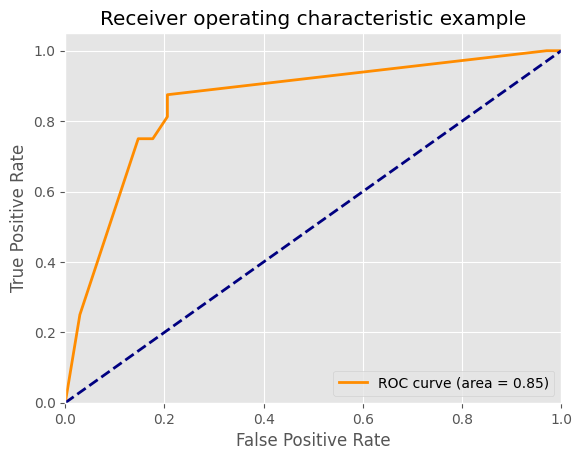
plt.plot(fpr[2], tpr[2], color='darkorange',
lw=lw, label='ROC curve (area = %0.2f)' % roc_auc[2])
plt.plot([0, 1], [0, 1], color='navy', lw=lw, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc="lower right")
plt.show()
整合學習案例1——幸福感預測
背景介紹
此案例是一個資料探勘型別的比賽——幸福感預測的baseline。比賽的資料使用的是官方的《中國綜合社會調查(CGSS)》檔案中的調查結果中的資料,其共包含有139個維度的特徵,包括個體變數(性別、年齡、地域、職業、健康、婚姻與政治面貌等等)、家庭變數(父母、配偶、子女、家庭資本等等)、社會態度(公平、信用、公共服務)等特徵。賽題要求使用以上 139 維的特徵,使用 8000 餘組資料進行對於個人幸福感的預測(預測值為1,2,3,4,5,其中1代表幸福感最低,5代表幸福感最高)
具體程式碼
匯入需要的包
import os
import time
import pandas as pd
import numpy as np
import seaborn as sns
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn import metrics
from datetime import datetime
import matplotlib.pyplot as plt
from sklearn.metrics import roc_auc_score, roc_curve, mean_squared_error,mean_absolute_error, f1_score
import lightgbm as lgb
import xgboost as xgb
from sklearn.ensemble import RandomForestRegressor as rfr
from sklearn.ensemble import ExtraTreesRegressor as etr
from sklearn.linear_model import BayesianRidge as br
from sklearn.ensemble import GradientBoostingRegressor as gbr
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.linear_model import LinearRegression as lr
from sklearn.linear_model import ElasticNet as en
from sklearn.kernel_ridge import KernelRidge as kr
from sklearn.model_selection import KFold, StratifiedKFold,GroupKFold, RepeatedKFold
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn import preprocessing
import logging
import warnings
warnings.filterwarnings('ignore') #消除warning
匯入資料集
train = pd.read_csv("train.csv", parse_dates=['survey_time'],encoding='latin-1')
# 第二個引數代表把那一列轉換為時間型別
test = pd.read_csv("test.csv", parse_dates=['survey_time'],encoding='latin-1')
#latin-1向下相容ASCII
train = train[train["happiness"] != -8].reset_index(drop=True)
# =8代表沒有回答是否幸福因此不要這些,reset_index是重置索引,因為行數變化了
train_data_copy = train.copy() # 完全刪除掉-8的行
target_col = "happiness"
target = train_data_copy[target_col]
del train_data_copy[target_col]
data = pd.concat([train_data_copy, test], axis = 0, ignore_index = True)
# 把他們按照行疊在一起
資料預處理
資料中出現的主要負數值是-1,-2,-3,-8,因此分別定義函數可以處理。
#make feature +5
#csv中有複數值:-1、-2、-3、-8,將他們視為有問題的特徵,但是不刪去
def getres1(row):
return len([x for x in row.values if type(x)==int and x<0])
def getres2(row):
return len([x for x in row.values if type(x)==int and x==-8])
def getres3(row):
return len([x for x in row.values if type(x)==int and x==-1])
def getres4(row):
return len([x for x in row.values if type(x)==int and x==-2])
def getres5(row):
return len([x for x in row.values if type(x)==int and x==-3])
#檢查資料
# 這裡的意思就是將函數應用到表格中,而axis=1是應用到每一行,因此得到新的特徵行數跟原來
# 是一樣的,統計了該行為負數的個數
data['neg1'] = data[data.columns].apply(lambda row:getres1(row),axis=1)
data.loc[data['neg1']>20,'neg1'] = 20 #平滑處理
data['neg2'] = data[data.columns].apply(lambda row:getres2(row),axis=1)
data['neg3'] = data[data.columns].apply(lambda row:getres3(row),axis=1)
data['neg4'] = data[data.columns].apply(lambda row:getres4(row),axis=1)
data['neg5'] = data[data.columns].apply(lambda row:getres5(row),axis=1)
而對於缺失值的填補,需要針對特徵加上自己的理解,例如如果家庭成員缺失值,那麼就填補為1,例如家庭收入確實,那麼可以填補為整個特徵的平均值等等,這部分可以自己發揮想象力,採用fillna進行填補。
先檢視哪些列存在缺失值需要填補:
data.isnull().sum()[data.isnull().sum() > 0]
edu_other 10950
edu_status 1569
edu_yr 2754
join_party 9831
property_other 10867
hukou_loc 4
social_neighbor 1096
social_friend 1096
work_status 6932
work_yr 6932
work_type 6931
work_manage 6931
family_income 1
invest_other 10911
minor_child 1447
marital_1st 1128
s_birth 2365
marital_now 2445
s_edu 2365
s_political 2365
s_hukou 2365
s_income 2365
s_work_exper 2365
s_work_status 7437
s_work_type 7437
dtype: int64
那麼對以上這些特徵進行處理:
data['work_status'] = data['work_status'].fillna(0)
data['work_yr'] = data['work_yr'].fillna(0)
data['work_manage'] = data['work_manage'].fillna(0)
data['work_type'] = data['work_type'].fillna(0)
data['edu_yr'] = data['edu_yr'].fillna(0)
data['edu_status'] = data['edu_status'].fillna(0)
data['s_work_type'] = data['s_work_type'].fillna(0)
data['s_work_status'] = data['s_work_status'].fillna(0)
data['s_political'] = data['s_political'].fillna(0)
data['s_hukou'] = data['s_hukou'].fillna(0)
data['s_income'] = data['s_income'].fillna(0)
data['s_birth'] = data['s_birth'].fillna(0)
data['s_edu'] = data['s_edu'].fillna(0)
data['s_work_exper'] = data['s_work_exper'].fillna(0)
data['minor_child'] = data['minor_child'].fillna(0)
data['marital_now'] = data['marital_now'].fillna(0)
data['marital_1st'] = data['marital_1st'].fillna(0)
data['social_neighbor']=data['social_neighbor'].fillna(0)
data['social_friend']=data['social_friend'].fillna(0)
data['hukou_loc']=data['hukou_loc'].fillna(1) #最少為1,表示戶口
data['family_income']=data['family_income'].fillna(66365) #刪除問題值後的平均值
特殊格式的資訊也需要處理,例如跟時間有關的資訊,可以化成對應方便處理的格式,以及對年齡進行分段處理:
data['survey_time'] = pd.to_datetime(data['survey_time'], format='%Y-%m-%d',
errors='coerce')
#防止時間格式不同的報錯errors='coerce‘,格式不對就幅值NaN
data['survey_time'] = data['survey_time'].dt.year #僅僅是year,方便計算年齡
data['age'] = data['survey_time']-data['birth']
bins = [0,17,26,34,50,63,100]
data['age_bin'] = pd.cut(data['age'], bins, labels=[0,1,2,3,4,5])
還有就是對一些異常值的處理,例如在某些問題中不應該出現負數但是出現了負數,那麼就可以根據我們的直觀理解來進行處理:
# 對宗教進行處理
data.loc[data['religion'] < 0, 'religion'] = 1 # 1代表不信仰宗教
data.loc[data['religion_freq'] < 0, "religion_freq"] = 0 # 代表不參加
# 教育
data.loc[data['edu'] < 0, 'edu'] = 1 # 負數說明沒有接受過任何教育
data.loc[data['edu_status'] < 0 , 'edu_status'] = 0
data.loc[data['edu_yr']<0,'edu_yr'] = 0
# 收入
data.loc[data['income']<0,'income'] = 0 #認為無收入
#對‘政治面貌’處理
data.loc[data['political']<0,'political'] = 1 #認為是群眾
#對體重處理
data.loc[(data['weight_jin']<=80)&(data['height_cm']>=160),'weight_jin']= data['weight_jin']*2
data.loc[data['weight_jin']<=60,'weight_jin']= data['weight_jin']*2 #沒有60斤的成年人吧
#對身高處理
data.loc[data['height_cm']<130,'height_cm'] = 150 #成年人的實際情況
#對‘健康’處理
data.loc[data['health']<0,'health'] = 4 #認為是比較健康
data.loc[data['health_problem']<0,'health_problem'] = 4 # 4代表很少
#對‘沮喪’處理
data.loc[data['depression']<0,'depression'] = 4 #很少
#對‘媒體’處理
data.loc[data['media_1']<0,'media_1'] = 1 #都是從不
data.loc[data['media_2']<0,'media_2'] = 1
data.loc[data['media_3']<0,'media_3'] = 1
data.loc[data['media_4']<0,'media_4'] = 1
data.loc[data['media_5']<0,'media_5'] = 1
data.loc[data['media_6']<0,'media_6'] = 1
#對‘空閒活動’處理
data.loc[data['leisure_1']<0,'leisure_1'] = data['leisure_1'].mode() # 眾數
data.loc[data['leisure_2']<0,'leisure_2'] = data['leisure_2'].mode()
data.loc[data['leisure_3']<0,'leisure_3'] = data['leisure_3'].mode()
data.loc[data['leisure_4']<0,'leisure_4'] = data['leisure_4'].mode()
data.loc[data['leisure_5']<0,'leisure_5'] = data['leisure_5'].mode()
data.loc[data['leisure_6']<0,'leisure_6'] = data['leisure_6'].mode()
data.loc[data['leisure_7']<0,'leisure_7'] = data['leisure_7'].mode()
data.loc[data['leisure_8']<0,'leisure_8'] = data['leisure_8'].mode()
data.loc[data['leisure_9']<0,'leisure_9'] = data['leisure_9'].mode()
data.loc[data['leisure_10']<0,'leisure_10'] = data['leisure_10'].mode()
data.loc[data['leisure_11']<0,'leisure_11'] = data['leisure_11'].mode()
data.loc[data['leisure_12']<0,'leisure_12'] = data['leisure_12'].mode()
data.loc[data['socialize']<0, 'socialize'] = 2 # 社交,很少
data.loc[data['relax']<0, 'relax'] = 2 # 放鬆,很少
data.loc[data['learn']<0, 'learn'] = 2 # 學習,很少
data.loc[data['social_neighbor']<0, 'social_neighbor'] = 6 # 一年1次或更少社交
data.loc[data['social_friend']<0, 'social_friend'] = 6
data.loc[data['socia_outing']<0, 'socia_outing'] = 1
data.loc[data['neighbor_familiarity']<0,'social_neighbor']= 2 # 不太熟悉
data.loc[data['equity']<0,'equity'] = 3 # 不知道公不公平
#對‘社會等級’處理
data.loc[data['class_10_before']<0,'class_10_before'] = 3
data.loc[data['class']<0,'class'] = 5
data.loc[data['class_10_after']<0,'class_10_after'] = 5
data.loc[data['class_14']<0,'class_14'] = 2
#對‘工作情況’處理
data.loc[data['work_status']<0,'work_status'] = 0
data.loc[data['work_yr']<0,'work_yr'] = 0
data.loc[data['work_manage']<0,'work_manage'] = 0
data.loc[data['work_type']<0,'work_type'] = 0
#對‘社會保障’處理
data.loc[data['insur_1']<0,'insur_1'] = 1
data.loc[data['insur_2']<0,'insur_2'] = 1
data.loc[data['insur_3']<0,'insur_3'] = 1
data.loc[data['insur_4']<0,'insur_4'] = 1
data.loc[data['insur_1']==0,'insur_1'] = 0
data.loc[data['insur_2']==0,'insur_2'] = 0
data.loc[data['insur_3']==0,'insur_3'] = 0
data.loc[data['insur_4']==0,'insur_4'] = 0
#對家庭情況處理
family_income_mean = data['family_income'].mean() # 計算均值
data.loc[data['family_income']<0,'family_income'] = family_income_mean
data.loc[data['family_m']<0,'family_m'] = 2
data.loc[data['family_status']<0,'family_status'] = 3
data.loc[data['house']<0,'house'] = 1
data.loc[data['car']<0,'car'] = 0
data.loc[data['car']==2,'car'] = 0 #變為0和1
data.loc[data['son']<0,'son'] = 0
data.loc[data['daughter']<0,'daughter'] = 0
data.loc[data['minor_child']<0,'minor_child'] = 0
#對‘婚姻’處理
data.loc[data['marital_1st']<0,'marital_1st'] = 0
data.loc[data['marital_now']<0,'marital_now'] = 0
#對‘配偶’處理
data.loc[data['s_birth']<0,'s_birth'] = 0
data.loc[data['s_edu']<0,'s_edu'] = 0
data.loc[data['s_political']<0,'s_political'] = 0
data.loc[data['s_hukou']<0,'s_hukou'] = 0
data.loc[data['s_income']<0,'s_income'] = 0
data.loc[data['s_work_type']<0,'s_work_type'] = 0
data.loc[data['s_work_status']<0,'s_work_status'] = 0
data.loc[data['s_work_exper']<0,'s_work_exper'] = 0
#對‘父母情況’處理
data.loc[data['f_birth']<0,'f_birth'] = 1945
data.loc[data['f_edu']<0,'f_edu'] = 1
data.loc[data['f_political']<0,'f_political'] = 1
data.loc[data['f_work_14']<0,'f_work_14'] = 2
data.loc[data['m_birth']<0,'m_birth'] = 1940
data.loc[data['m_edu']<0,'m_edu'] = 1
data.loc[data['m_political']<0,'m_political'] = 1
data.loc[data['m_work_14']<0,'m_work_14'] = 2
#和同齡人相比社會經濟地位
data.loc[data['status_peer']<0,'status_peer'] = 2
#和3年前比社會經濟地位
data.loc[data['status_3_before']<0,'status_3_before'] = 2
#對‘觀點’處理
data.loc[data['view']<0,'view'] = 4
#對期望年收入處理
data.loc[data['inc_ability']<=0,'inc_ability']= 2
inc_exp_mean = data['inc_exp'].mean()
data.loc[data['inc_exp']<=0,'inc_exp']= inc_exp_mean #取均值
#部分特徵處理,取眾數(首先去除缺失值的資料)
for i in range(1,9+1):
data.loc[data['public_service_'+str(i)]<0,'public_service_'+str(i)] = int(data['public_service_'+str(i)].dropna().mode().values)
for i in range(1,13+1):
data.loc[data['trust_'+str(i)]<0,'trust_'+str(i)] = int(data['trust_'+str(i)].dropna().mode().values)
#上面的迴圈要加上int,因為values返回一個array是1*1的,那麼跟右邊的長度匹配不上
# 轉換成int就可以廣播賦值
資料增廣
上述只是對原始的特徵進行處理,下面我們需要進一步分析特徵的關係,引入更多有可能具有較大重要性的特徵來協助判斷。
#第一次結婚年齡 147
data['marital_1stbir'] = data['marital_1st'] - data['birth']
#最近結婚年齡 148
data['marital_nowtbir'] = data['marital_now'] - data['birth']
#是否再婚 149
data['mar'] = data['marital_nowtbir'] - data['marital_1stbir']
#配偶年齡 150
data['marital_sbir'] = data['marital_now']-data['s_birth']
#配偶年齡差 151
data['age_'] = data['marital_nowtbir'] - data['marital_sbir']
#收入比 151+7 =158
data['income/s_income'] = data['income']/(data['s_income']+1) #同居伴侶
data['income+s_income'] = data['income']+(data['s_income']+1)
data['income/family_income'] = data['income']/(data['family_income']+1)
data['all_income/family_income'] = (data['income']+data['s_income'])/(data['family_income']+1)
data['income/inc_exp'] = data['income']/(data['inc_exp']+1)
data['family_income/m'] = data['family_income']/(data['family_m']+0.01)
data['income/m'] = data['income']/(data['family_m']+0.01)
#收入/面積比 158+4=162
data['income/floor_area'] = data['income']/(data['floor_area']+0.01)
data['all_income/floor_area'] = (data['income']+data['s_income'])/(data['floor_area']+0.01)
data['family_income/floor_area'] = data['family_income']/(data['floor_area']+0.01)
data['floor_area/m'] = data['floor_area']/(data['family_m']+0.01)
#class 162+3=165
data['class_10_diff'] = (data['class_10_after'] - data['class'])
data['class_diff'] = data['class'] - data['class_10_before']
data['class_14_diff'] = data['class'] - data['class_14']
#悠閒指數 166
leisure_fea_lis = ['leisure_'+str(i) for i in range(1,13)]
data['leisure_sum'] = data[leisure_fea_lis].sum(axis=1) #skew
#滿意指數 167
public_service_fea_lis = ['public_service_'+str(i) for i in range(1,10)]
data['public_service_sum'] = data[public_service_fea_lis].sum(axis=1) #skew
#信任指數 168
trust_fea_lis = ['trust_'+str(i) for i in range(1,14)]
data['trust_sum'] = data[trust_fea_lis].sum(axis=1) #skew
#province mean 168+13=181
data['province_income_mean'] = data.groupby(['province'])['income'].transform('mean').values
data['province_family_income_mean'] = data.groupby(['province'])['family_income'].transform('mean').values
data['province_equity_mean'] = data.groupby(['province'])['equity'].transform('mean').values
data['province_depression_mean'] = data.groupby(['province'])['depression'].transform('mean').values
data['province_floor_area_mean'] = data.groupby(['province'])['floor_area'].transform('mean').values
data['province_health_mean'] = data.groupby(['province'])['health'].transform('mean').values
data['province_class_10_diff_mean'] = data.groupby(['province'])['class_10_diff'].transform('mean').values
data['province_class_mean'] = data.groupby(['province'])['class'].transform('mean').values
data['province_health_problem_mean'] = data.groupby(['province'])['health_problem'].transform('mean').values
data['province_family_status_mean'] = data.groupby(['province'])['family_status'].transform('mean').values
data['province_leisure_sum_mean'] = data.groupby(['province'])['leisure_sum'].transform('mean').values
data['province_public_service_sum_mean'] = data.groupby(['province'])['public_service_sum'].transform('mean').values
data['province_trust_sum_mean'] = data.groupby(['province'])['trust_sum'].transform('mean').values
#city mean 181+13=194
data['city_income_mean'] = data.groupby(['city'])['income'].transform('mean').values #按照city分組
data['city_family_income_mean'] = data.groupby(['city'])['family_income'].transform('mean').values
data['city_equity_mean'] = data.groupby(['city'])['equity'].transform('mean').values
data['city_depression_mean'] = data.groupby(['city'])['depression'].transform('mean').values
data['city_floor_area_mean'] = data.groupby(['city'])['floor_area'].transform('mean').values
data['city_health_mean'] = data.groupby(['city'])['health'].transform('mean').values
data['city_class_10_diff_mean'] = data.groupby(['city'])['class_10_diff'].transform('mean').values
data['city_class_mean'] = data.groupby(['city'])['class'].transform('mean').values
data['city_health_problem_mean'] = data.groupby(['city'])['health_problem'].transform('mean').values
data['city_family_status_mean'] = data.groupby(['city'])['family_status'].transform('mean').values
data['city_leisure_sum_mean'] = data.groupby(['city'])['leisure_sum'].transform('mean').values
data['city_public_service_sum_mean'] = data.groupby(['city'])['public_service_sum'].transform('mean').values
data['city_trust_sum_mean'] = data.groupby(['city'])['trust_sum'].transform('mean').values
#county mean 194 + 13 = 207
data['county_income_mean'] = data.groupby(['county'])['income'].transform('mean').values
data['county_family_income_mean'] = data.groupby(['county'])['family_income'].transform('mean').values
data['county_equity_mean'] = data.groupby(['county'])['equity'].transform('mean').values
data['county_depression_mean'] = data.groupby(['county'])['depression'].transform('mean').values
data['county_floor_area_mean'] = data.groupby(['county'])['floor_area'].transform('mean').values
data['county_health_mean'] = data.groupby(['county'])['health'].transform('mean').values
data['county_class_10_diff_mean'] = data.groupby(['county'])['class_10_diff'].transform('mean').values
data['county_class_mean'] = data.groupby(['county'])['class'].transform('mean').values
data['county_health_problem_mean'] = data.groupby(['county'])['health_problem'].transform('mean').values
data['county_family_status_mean'] = data.groupby(['county'])['family_status'].transform('mean').values
data['county_leisure_sum_mean'] = data.groupby(['county'])['leisure_sum'].transform('mean').values
data['county_public_service_sum_mean'] = data.groupby(['county'])['public_service_sum'].transform('mean').values
data['county_trust_sum_mean'] = data.groupby(['county'])['trust_sum'].transform('mean').values
#ratio 相比同省 207 + 13 =220
data['income/province'] = data['income']/(data['province_income_mean'])
data['family_income/province'] = data['family_income']/(data['province_family_income_mean'])
data['equity/province'] = data['equity']/(data['province_equity_mean'])
data['depression/province'] = data['depression']/(data['province_depression_mean'])
data['floor_area/province'] = data['floor_area']/(data['province_floor_area_mean'])
data['health/province'] = data['health']/(data['province_health_mean'])
data['class_10_diff/province'] = data['class_10_diff']/(data['province_class_10_diff_mean'])
data['class/province'] = data['class']/(data['province_class_mean'])
data['health_problem/province'] = data['health_problem']/(data['province_health_problem_mean'])
data['family_status/province'] = data['family_status']/(data['province_family_status_mean'])
data['leisure_sum/province'] = data['leisure_sum']/(data['province_leisure_sum_mean'])
data['public_service_sum/province'] = data['public_service_sum']/(data['province_public_service_sum_mean'])
data['trust_sum/province'] = data['trust_sum']/(data['province_trust_sum_mean']+1)
#ratio 相比同市 220 + 13 =233
data['income/city'] = data['income']/(data['city_income_mean'])
data['family_income/city'] = data['family_income']/(data['city_family_income_mean'])
data['equity/city'] = data['equity']/(data['city_equity_mean'])
data['depression/city'] = data['depression']/(data['city_depression_mean'])
data['floor_area/city'] = data['floor_area']/(data['city_floor_area_mean'])
data['health/city'] = data['health']/(data['city_health_mean'])
data['class_10_diff/city'] = data['class_10_diff']/(data['city_class_10_diff_mean'])
data['class/city'] = data['class']/(data['city_class_mean'])
data['health_problem/city'] = data['health_problem']/(data['city_health_problem_mean'])
data['family_status/city'] = data['family_status']/(data['city_family_status_mean'])
data['leisure_sum/city'] = data['leisure_sum']/(data['city_leisure_sum_mean'])
data['public_service_sum/city'] = data['public_service_sum']/(data['city_public_service_sum_mean'])
data['trust_sum/city'] = data['trust_sum']/(data['city_trust_sum_mean'])
#ratio 相比同個地區 233 + 13 =246
data['income/county'] = data['income']/(data['county_income_mean'])
data['family_income/county'] = data['family_income']/(data['county_family_income_mean'])
data['equity/county'] = data['equity']/(data['county_equity_mean'])
data['depression/county'] = data['depression']/(data['county_depression_mean'])
data['floor_area/county'] = data['floor_area']/(data['county_floor_area_mean'])
data['health/county'] = data['health']/(data['county_health_mean'])
data['class_10_diff/county'] = data['class_10_diff']/(data['county_class_10_diff_mean'])
data['class/county'] = data['class']/(data['county_class_mean'])
data['health_problem/county'] = data['health_problem']/(data['county_health_problem_mean'])
data['family_status/county'] = data['family_status']/(data['county_family_status_mean'])
data['leisure_sum/county'] = data['leisure_sum']/(data['county_leisure_sum_mean'])
data['public_service_sum/county'] = data['public_service_sum']/(data['county_public_service_sum_mean'])
data['trust_sum/county'] = data['trust_sum']/(data['county_trust_sum_mean'])
#age mean 246+ 13 =259
data['age_income_mean'] = data.groupby(['age'])['income'].transform('mean').values
data['age_family_income_mean'] = data.groupby(['age'])['family_income'].transform('mean').values
data['age_equity_mean'] = data.groupby(['age'])['equity'].transform('mean').values
data['age_depression_mean'] = data.groupby(['age'])['depression'].transform('mean').values
data['age_floor_area_mean'] = data.groupby(['age'])['floor_area'].transform('mean').values
data['age_health_mean'] = data.groupby(['age'])['health'].transform('mean').values
data['age_class_10_diff_mean'] = data.groupby(['age'])['class_10_diff'].transform('mean').values
data['age_class_mean'] = data.groupby(['age'])['class'].transform('mean').values
data['age_health_problem_mean'] = data.groupby(['age'])['health_problem'].transform('mean').values
data['age_family_status_mean'] = data.groupby(['age'])['family_status'].transform('mean').values
data['age_leisure_sum_mean'] = data.groupby(['age'])['leisure_sum'].transform('mean').values
data['age_public_service_sum_mean'] = data.groupby(['age'])['public_service_sum'].transform('mean').values
data['age_trust_sum_mean'] = data.groupby(['age'])['trust_sum'].transform('mean').values
# 和同齡人相比259 + 13 =272
data['income/age'] = data['income']/(data['age_income_mean'])
data['family_income/age'] = data['family_income']/(data['age_family_income_mean'])
data['equity/age'] = data['equity']/(data['age_equity_mean'])
data['depression/age'] = data['depression']/(data['age_depression_mean'])
data['floor_area/age'] = data['floor_area']/(data['age_floor_area_mean'])
data['health/age'] = data['health']/(data['age_health_mean'])
data['class_10_diff/age'] = data['class_10_diff']/(data['age_class_10_diff_mean'])
data['class/age'] = data['class']/(data['age_class_mean'])
data['health_problem/age'] = data['health_problem']/(data['age_health_problem_mean'])
data['family_status/age'] = data['family_status']/(data['age_family_status_mean'])
data['leisure_sum/age'] = data['leisure_sum']/(data['age_leisure_sum_mean'])
data['public_service_sum/age'] = data['public_service_sum']/(data['age_public_service_sum_mean'])
data['trust_sum/age'] = data['trust_sum']/(data['age_trust_sum_mean'])
經過上述的處理,特徵擴充為272維,而經過分析發現應該刪除掉有效樣本數過少的特徵:
#刪除數值特別少的和之前用過的特徵
del_list=['id','survey_time','edu_other','invest_other','property_other','join_party','province','city','county']
use_feature = [clo for clo in data.columns if clo not in del_list]
data.fillna(0,inplace=True) #還是補0
train_shape = train.shape[0] #一共的資料量,訓練集
features = data[use_feature].columns #刪除後所有的特徵
X_train_263 = data[:train_shape][use_feature].values
y_train = target
X_test_263 = data[train_shape:][use_feature].values
X_train_263.shape #最終一種263個特徵
再結合個人經驗,挑選出最重要的49個特徵:
imp_fea_49 = ['equity','depression','health','class','family_status','health_problem','class_10_after',
'equity/province','equity/city','equity/county',
'depression/province','depression/city','depression/county',
'health/province','health/city','health/county',
'class/province','class/city','class/county',
'family_status/province','family_status/city','family_status/county',
'family_income/province','family_income/city','family_income/county',
'floor_area/province','floor_area/city','floor_area/county',
'leisure_sum/province','leisure_sum/city','leisure_sum/county',
'public_service_sum/province','public_service_sum/city','public_service_sum/county',
'trust_sum/province','trust_sum/city','trust_sum/county',
'income/m','public_service_sum','class_diff','status_3_before','age_income_mean','age_floor_area_mean',
'weight_jin','height_cm',
'health/age','depression/age','equity/age','leisure_sum/age'
]
train_shape = train.shape[0]
X_train_49 = data[:train_shape][imp_fea_49].values
X_test_49 = data[train_shape:][imp_fea_49].values
# X_train_49.shape #最重要的49個特徵
再將其中一些特徵轉換為one-hot特徵:
from sklearn import preprocessing
cat_fea = ['survey_type','gender','nationality','edu_status','political','hukou','hukou_loc','work_exper','work_status','work_type',
'work_manage','marital','s_political','s_hukou','s_work_exper','s_work_status','s_work_type','f_political','f_work_14',
'm_political','m_work_14'] #已經是0、1的值不需要onehot
noc_fea = [clo for clo in use_feature if clo not in cat_fea]
onehot_data = data[cat_fea].values
enc = preprocessing.OneHotEncoder(categories = 'auto')
oh_data=enc.fit_transform(onehot_data).toarray()
oh_data.shape #變為onehot編碼格式
X_train_oh = oh_data[:train_shape,:]
X_test_oh = oh_data[train_shape:,:]
X_train_oh.shape #其中的訓練集
X_train_383 = np.column_stack([data[:train_shape][noc_fea].values,X_train_oh])#先是noc,再是cat_fea
X_test_383 = np.column_stack([data[train_shape:][noc_fea].values,X_test_oh])
# X_train_383.shape
最終得到為383個特徵。
模型構建
此處模型構建的主要流程為:
- 構建多個整合分類演演算法進行訓練,並對驗證集進行預測
- 將各個基礎整合分類演演算法對驗證集的預測作為訓練資料,驗證集的標籤作為訓練標籤,由此來訓練一個迴歸模型對各個基礎整合分類演演算法進行融合,得到較好的結果
具體處理的流程是建立一個模型,然後對其採用五折交叉驗證的方式去訓練,並且對驗證集和測試集進行預測,多個模型得到的驗證集預測進行堆疊得到下一層的訓練資料,而多個模型得到的測試集預測直接進行平均求和,以此給訓練好的下一層做出真正對測試集的預測。
對原始263維特徵進行處理
LightGBM
lgb_263_param = { # 這是訓練的參數列
"num_leaves":7,
"min_data_in_leaf": 20, # 一個葉子上最小分配到的數量,用來處理過擬合
"objective": "regression", # 設定型別為迴歸
"max_depth": -1, # 限制樹的最大深度,-1代表沒有限制
"learning_rate": 0.003,
"boosting": "gbdt", # 用gbdt演演算法
"feature_fraction": 0.18, # 每次迭代時使用18%的特徵參與建樹,引入特徵子空間的多樣性
"bagging_freq": 1, # 每一次迭代都執行bagging
"bagging_fraction": 0.55, # 每次bagging在不進行重取樣的情況下隨機選擇55%資料訓練
"bagging_seed": 14,
"metric": 'mse',
"lambda_l1": 0.1,
"lambda_l2": 0.2,
"verbosity": -1 # 列印訊息的詳細程度
}
folds = StratifiedKFold(n_splits=5, shuffle=True, random_state = 4)
# 產生一個容器,可以用來對對資料集進行打亂的5次切分,以此來進行五折交叉驗證
oof_lgb_263 = np.zeros(len(X_train_263))
predictions_lgb_263 = np.zeros(len(X_test_263))
for fold_, (train_idx, valid_idx) in enumerate(folds.split(X_train_263, y_train)):
# 切分後返回的訓練集和驗證集的索引
print("fold n{}".format(fold_+1)) # 當前第幾折
train_data_now = lgb.Dataset(X_train_263[train_idx], y_train[train_idx])
valid_data_now = lgb.Dataset(X_train_263[valid_idx], y_train[valid_idx])
# 取出資料並轉換為lgb的資料
num_round = 10000
lgb_263 = lgb.train(lgb_263_param, train_data_now, num_round,
valid_sets=[train_data_now, valid_data_now], verbose_eval=500,
early_stopping_rounds = 800)
# 第一個引數是原始引數,第二個是用來訓練的資料,第三個引數代表迭代次數
# 第四個引數是訓練後評估所用的資料,第五個引數是每多少次迭代就列印下當前的評估資訊
# 第六個引數是超過多少次迭代沒有變化就停止
oof_lgb_263[valid_idx] = lgb_263.predict(X_train_263[valid_idx],
num_iteration=lgb_263.best_iteration)
predictions_lgb_263 += lgb_263.predict(X_test_263, num_iteration=
lgb_263.best_iteration) / folds.n_splits
# 這是將預測概率進行平均
print("CV score: {:<8.8f}".format(mean_squared_error(oof_lgb_263, target)))
# 它是迴歸型別因此不能用正確率來衡量
由於這次列印出來的資訊過多,因此我只補充最終的分數:
CV score: 0.45153757
而我們可以利用lightGBM中的特徵重要性來直觀展現各個特徵的重要性程度:
pd.set_option("display.max_columns", None) # 設定可以顯示的最大行和最大列
pd.set_option('display.max_rows', None) # 如果超過就顯示省略號,none表示不省略
#設定value的顯示長度為100,預設為50
pd.set_option('max_colwidth',100)
# 建立,然後只有一列就是剛才所使用的的特徵
df = pd.DataFrame(data[use_feature].columns.tolist(), columns=['feature'])
df['importance'] = list(lgb_263.feature_importance())
df = df.sort_values(by='importance', ascending=False) # 降序排列
plt.figure(figsize = (14,28))
sns.barplot(x='importance', y='feature', data = df.head(50))# 取出前五十個畫圖
plt.title('Features importance (averaged/folds)')
plt.tight_layout() # 自動調整適應範圍
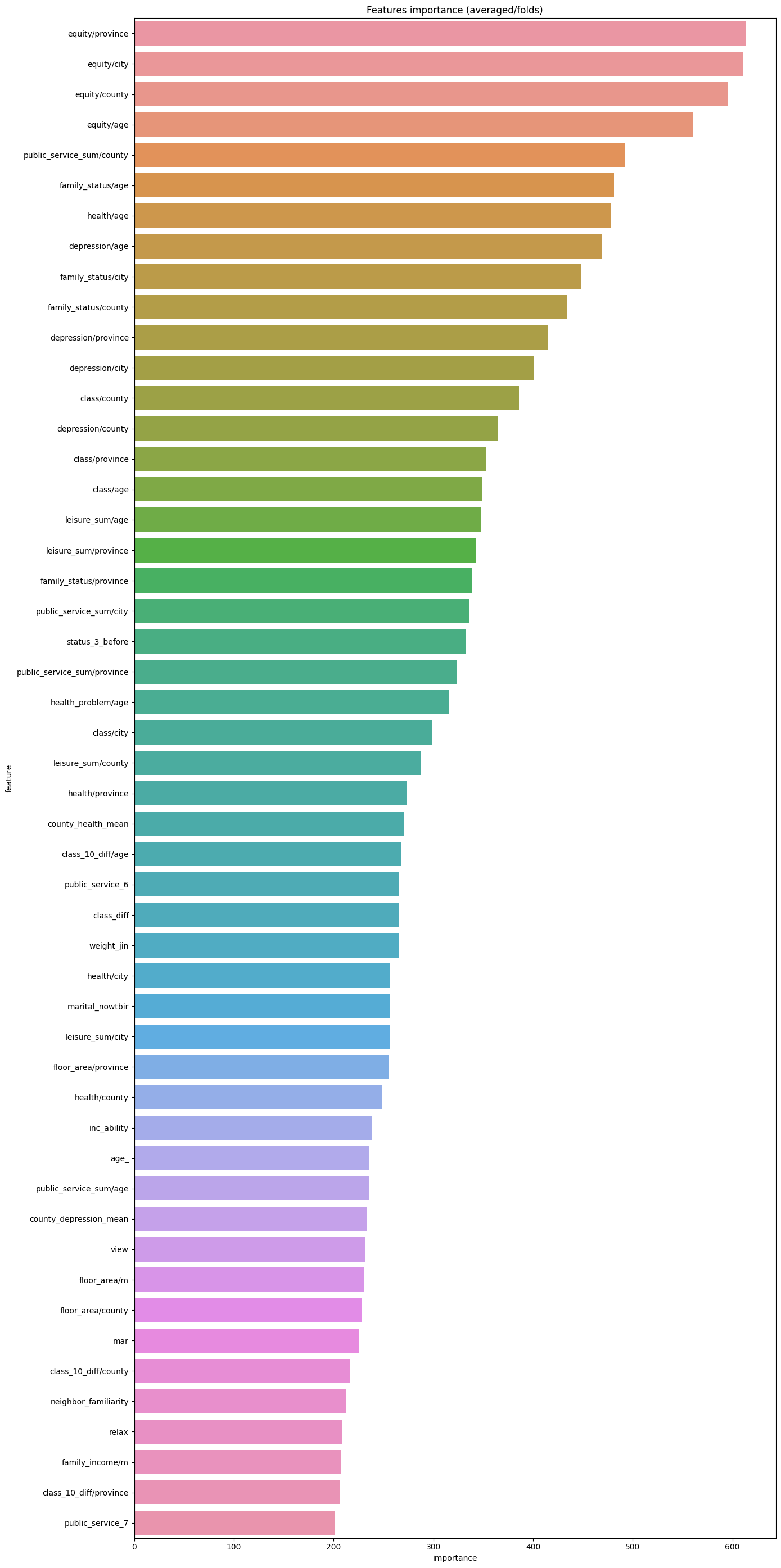
可以看到最重要的特徵為同齡人的健康程度,這方面還是很符合直觀感覺的。
xgBoost
xgb_263_params = {
"eta":0.02, # 學習率
"max_depth": 6, # 樹的最大深度
"min_child_weight":3, # 劃分為葉結點所含樣本的最小權重和
"gamma":0, # 在分裂時損失最小應該減少gamma,越大則越減少過擬合
"subsample":0.7, # 控制對於每棵樹隨機取樣的比例
"colsample_bytree":0.3, # 用來控制每棵樹對於特徵的取樣佔比
"lambda":2,
"objective":"reg:linear",
"eval_metric":"rmse",
"verbosity":1, # 列印訊息的詳細程度
"nthread":-1 # 並行執行緒數,使用最大
}
folds = StratifiedKFold(n_splits=5, shuffle=True, random_state=2019)
oof_xgb_263 = np.zeros(len(X_train_263))
predictions_xgb_263 = np.zeros(len(X_test_263))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_263, y_train)):
print("fold n°{}".format(fold_+1))
trn_data = xgb.DMatrix(X_train_263[trn_idx], y_train[trn_idx])
val_data = xgb.DMatrix(X_train_263[val_idx], y_train[val_idx])
watchlist = [(trn_data, 'train'), (val_data, 'valid_data')]
xgb_263 = xgb.train(dtrain=trn_data, num_boost_round=3000, evals=watchlist,
early_stopping_rounds=600, verbose_eval=500,
params=xgb_263_params)
oof_xgb_263[val_idx] = xgb_263.predict(xgb.DMatrix(X_train_263[val_idx]),
ntree_limit=xgb_263.best_ntree_limit)
predictions_xgb_263 += xgb_263.predict(xgb.DMatrix(X_test_263), ntree_limit=
xgb_263.best_ntree_limit) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_xgb_263, target)))
CV score: 0.45402538
隨機森林
#RandomForestRegressor隨機森林
folds = KFold(n_splits=5, shuffle=True, random_state=2019)
oof_rfr_263 = np.zeros(len(X_train_263))
predictions_rfr_263 = np.zeros(len(X_test_263))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_263, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_263[trn_idx]
tr_y = y_train[trn_idx]
rfr_263 = rfr(n_estimators=1600,max_depth=9, min_samples_leaf=9,
min_weight_fraction_leaf=0.0,max_features=0.25,
verbose=1,n_jobs=-1) #並行化
#verbose = 0 為不在標準輸出流輸出紀錄檔資訊
#verbose = 1 為輸出進度條記錄
#verbose = 2 為每個epoch輸出一行記錄
rfr_263.fit(tr_x,tr_y)
oof_rfr_263[val_idx] = rfr_263.predict(X_train_263[val_idx])
predictions_rfr_263 += rfr_263.predict(X_test_263) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_rfr_263, target)))
CV score: 0.47810816
梯度提升GBDT
#GradientBoostingRegressor梯度提升決策樹
folds = StratifiedKFold(n_splits=5, shuffle=True, random_state=2018)
oof_gbr_263 = np.zeros(train_shape)
predictions_gbr_263 = np.zeros(len(X_test_263))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_263, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_263[trn_idx]
tr_y = y_train[trn_idx]
gbr_263 = gbr(n_estimators=400, learning_rate=0.01,subsample=0.65,max_depth=7, min_samples_leaf=20,
max_features=0.22,verbose=1)
gbr_263.fit(tr_x,tr_y)
oof_gbr_263[val_idx] = gbr_263.predict(X_train_263[val_idx])
predictions_gbr_263 += gbr_263.predict(X_test_263) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_gbr_263, target)))
CV score: 0.45781356
極端隨機森林迴歸
#ExtraTreesRegressor 極端隨機森林迴歸
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_etr_263 = np.zeros(train_shape)
predictions_etr_263 = np.zeros(len(X_test_263))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_263, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_263[trn_idx]
tr_y = y_train[trn_idx]
etr_263 = etr(n_estimators=1000,max_depth=8, min_samples_leaf=12, min_weight_fraction_leaf=0.0,
max_features=0.4,verbose=1,n_jobs=-1)# max_feature:劃分時考慮的最大特徵數
etr_263.fit(tr_x,tr_y)
oof_etr_263[val_idx] = etr_263.predict(X_train_263[val_idx])
predictions_etr_263 += etr_263.predict(X_test_263) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_etr_263, target)))
CV score: 0.48598827
綜上,我們得到了五個模型的預測結果、模型架構以及引數。那麼我們需要對這五個模型進行融合,可以用一個Kernel Ridge Regression,核脊迴歸來進行融合,這個迴歸有點類似於嶺迴歸,也是採用五折交叉驗證重複兩次:
train_stack2 = np.vstack([oof_lgb_263,oof_xgb_263,oof_gbr_263,oof_rfr_263,oof_etr_263]).transpose()
# transpose()函數的作用就是調換x,y,z的位置,也就是陣列的索引值,變成zyx
# 本來是5*驗證集樣本數*1,變成1*驗證集樣本數*5,二維矩陣每一行是單個樣本在5個模型上的預測結果,行數是樣本數
test_stack2 = np.vstack([predictions_lgb_263, predictions_xgb_263,predictions_gbr_263,predictions_rfr_263,predictions_etr_263]).transpose()
# 變成1*測試集樣本數*5,二維矩陣每一行是單個測試集樣本在5個模型上的預測結果,行數是樣本數
#交叉驗證:5折,重複2次
folds_stack = RepeatedKFold(n_splits=5, n_repeats=2, random_state=7)
oof_stack2 = np.zeros(train_stack2.shape[0])
predictions_lr2 = np.zeros(test_stack2.shape[0])
for fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack2,target)):
print("fold {}".format(fold_))
trn_data, trn_y = train_stack2[trn_idx], target.iloc[trn_idx].values
val_data, val_y = train_stack2[val_idx], target.iloc[val_idx].values
#Kernel Ridge Regression
lr2 = kr()
lr2.fit(trn_data, trn_y)
oof_stack2[val_idx] = lr2.predict(val_data)
predictions_lr2 += lr2.predict(test_stack2) / 10
mean_squared_error(target.values, oof_stack2)
0.44815130114230267
對49維的特徵進行處理
同樣我們可以對49維度的特徵進行類似處理,我將程式碼總結如下:
##### lgb_49
lgb_49_param = {
'num_leaves': 9,
'min_data_in_leaf': 23,
'objective':'regression',
'max_depth': -1,
'learning_rate': 0.002,
"boosting": "gbdt",
"feature_fraction": 0.45,
"bagging_freq": 1,
"bagging_fraction": 0.65,
"bagging_seed": 15,
"metric": 'mse',
"lambda_l2": 0.2,
"verbosity": -1} # 一個葉子上資料的最小數量 \ feature_fraction將會在每棵樹訓練之前選擇 45% 的特徵。可以用來加速訓練,可以用來處理過擬合。 #bagging_fraction不進行重取樣的情況下隨機選擇部分資料。可以用來加速訓練,可以用來處理過擬合。
folds = StratifiedKFold(n_splits=5, shuffle=True, random_state=9)
oof_lgb_49 = np.zeros(len(X_train_49))
predictions_lgb_49 = np.zeros(len(X_test_49))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
trn_data = lgb.Dataset(X_train_49[trn_idx], y_train[trn_idx])
val_data = lgb.Dataset(X_train_49[val_idx], y_train[val_idx])
num_round = 12000
lgb_49 = lgb.train(lgb_49_param, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=1000, early_stopping_rounds = 1000)
oof_lgb_49[val_idx] = lgb_49.predict(X_train_49[val_idx], num_iteration=lgb_49.best_iteration)
predictions_lgb_49 += lgb_49.predict(X_test_49, num_iteration=lgb_49.best_iteration) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_lgb_49, target)))
##### xgb_49
xgb_49_params = {'eta': 0.02,
'max_depth': 5,
'min_child_weight':3,
'gamma':0,
'subsample': 0.7,
'colsample_bytree': 0.35,
'lambda':2,
'objective': 'reg:linear',
'eval_metric': 'rmse',
'silent': True,
'nthread': -1}
folds = KFold(n_splits=5, shuffle=True, random_state=2019)
oof_xgb_49 = np.zeros(len(X_train_49))
predictions_xgb_49 = np.zeros(len(X_test_49))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
trn_data = xgb.DMatrix(X_train_49[trn_idx], y_train[trn_idx])
val_data = xgb.DMatrix(X_train_49[val_idx], y_train[val_idx])
watchlist = [(trn_data, 'train'), (val_data, 'valid_data')]
xgb_49 = xgb.train(dtrain=trn_data, num_boost_round=3000, evals=watchlist, early_stopping_rounds=600, verbose_eval=500, params=xgb_49_params)
oof_xgb_49[val_idx] = xgb_49.predict(xgb.DMatrix(X_train_49[val_idx]), ntree_limit=xgb_49.best_ntree_limit)
predictions_xgb_49 += xgb_49.predict(xgb.DMatrix(X_test_49), ntree_limit=xgb_49.best_ntree_limit) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_xgb_49, target)))
folds = StratifiedKFold(n_splits=5, shuffle=True, random_state=2018)
oof_gbr_49 = np.zeros(train_shape)
predictions_gbr_49 = np.zeros(len(X_test_49))
#GradientBoostingRegressor梯度提升決策樹
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_49[trn_idx]
tr_y = y_train[trn_idx]
gbr_49 = gbr(n_estimators=600, learning_rate=0.01,subsample=0.65,max_depth=6, min_samples_leaf=20,
max_features=0.35,verbose=1)
gbr_49.fit(tr_x,tr_y)
oof_gbr_49[val_idx] = gbr_49.predict(X_train_49[val_idx])
predictions_gbr_49 += gbr_49.predict(X_test_49) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_gbr_49, target)))
這裡由於特徵數目較少,只考慮使用3個基礎模型,接下來便是進行融合:
train_stack3 = np.vstack([oof_lgb_49,oof_xgb_49,oof_gbr_49]).transpose()
test_stack3 = np.vstack([predictions_lgb_49, predictions_xgb_49,predictions_gbr_49]).transpose()
#
folds_stack = RepeatedKFold(n_splits=5, n_repeats=2, random_state=7)
oof_stack3 = np.zeros(train_stack3.shape[0])
predictions_lr3 = np.zeros(test_stack3.shape[0])
for fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack3,target)):
print("fold {}".format(fold_))
trn_data, trn_y = train_stack3[trn_idx], target.iloc[trn_idx].values
val_data, val_y = train_stack3[val_idx], target.iloc[val_idx].values
#Kernel Ridge Regression
lr3 = kr()
lr3.fit(trn_data, trn_y)
oof_stack3[val_idx] = lr3.predict(val_data)
predictions_lr3 += lr3.predict(test_stack3) / 10
mean_squared_error(target.values, oof_stack3)
0.4662728551415085
對383維的特徵進行處理
這裡對383維度的特徵採用剛才類似的方法是可以的,但是也可以採用其他的方法,例如我們將基模型換成普通的迴歸模型:
Kernel Ridge Regression 基於核的嶺迴歸
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_kr_383 = np.zeros(train_shape)
predictions_kr_383 = np.zeros(len(X_test_383))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_383, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_383[trn_idx]
tr_y = y_train[trn_idx]
#Kernel Ridge Regression 嶺迴歸
kr_383 = kr()
kr_383.fit(tr_x,tr_y)
oof_kr_383[val_idx] = kr_383.predict(X_train_383[val_idx])
predictions_kr_383 += kr_383.predict(X_test_383) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_kr_383, target)))
CV score: 0.51412085
可以看到普通迴歸的誤差相比於整合演演算法會高一些。
普通嶺迴歸
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_ridge_383 = np.zeros(train_shape)
predictions_ridge_383 = np.zeros(len(X_test_383))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_383, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_383[trn_idx]
tr_y = y_train[trn_idx]
#使用嶺迴歸
ridge_383 = Ridge(alpha=1200)
ridge_383.fit(tr_x,tr_y)
oof_ridge_383[val_idx] = ridge_383.predict(X_train_383[val_idx])
predictions_ridge_383 += ridge_383.predict(X_test_383) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_ridge_383, target)))
CV score: 0.48687670
ElasticNet 彈性網路
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_en_383 = np.zeros(train_shape)
predictions_en_383 = np.zeros(len(X_test_383))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_383, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_383[trn_idx]
tr_y = y_train[trn_idx]
#ElasticNet 彈性網路
en_383 = en(alpha=1.0,l1_ratio=0.06)
en_383.fit(tr_x,tr_y)
oof_en_383[val_idx] = en_383.predict(X_train_383[val_idx])
predictions_en_383 += en_383.predict(X_test_383) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_en_383, target)))
CV score: 0.53296555
BayesianRidge 貝葉斯嶺迴歸
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_br_383 = np.zeros(train_shape)
predictions_br_383 = np.zeros(len(X_test_383))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_383, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_383[trn_idx]
tr_y = y_train[trn_idx]
#BayesianRidge 貝葉斯迴歸
br_383 = br()
br_383.fit(tr_x,tr_y)
oof_br_383[val_idx] = br_383.predict(X_train_383[val_idx])
predictions_br_383 += br_383.predict(X_test_383) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_br_383, target)))
CV score: 0.48717310
那麼對383特徵的四次迴歸也進行融合:
train_stack1 = np.vstack([oof_br_383,oof_kr_383,oof_en_383,oof_ridge_383]).transpose()
test_stack1 = np.vstack([predictions_br_383, predictions_kr_383,predictions_en_383,predictions_ridge_383]).transpose()
folds_stack = RepeatedKFold(n_splits=5, n_repeats=2, random_state=7)
oof_stack1 = np.zeros(train_stack1.shape[0])
predictions_lr1 = np.zeros(test_stack1.shape[0])
for fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack1,target)):
print("fold {}".format(fold_))
trn_data, trn_y = train_stack1[trn_idx], target.iloc[trn_idx].values
val_data, val_y = train_stack1[val_idx], target.iloc[val_idx].values
# LinearRegression簡單的線性迴歸
lr1 = lr()
lr1.fit(trn_data, trn_y)
oof_stack1[val_idx] = lr1.predict(val_data)
predictions_lr1 += lr1.predict(test_stack1) / 10
mean_squared_error(target.values, oof_stack1)
0.4878202780283125
對49維特徵的繼續處理
由於49維的特徵是最重要的特徵,所以這裡考慮增加更多的模型進行49維特徵的資料的構建工作。加入跟383特徵一樣的迴歸模型,具體如下:
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_kr_49 = np.zeros(train_shape)
predictions_kr_49 = np.zeros(len(X_test_49))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_49[trn_idx]
tr_y = y_train[trn_idx]
kr_49 = kr()
kr_49.fit(tr_x,tr_y)
oof_kr_49[val_idx] = kr_49.predict(X_train_49[val_idx])
predictions_kr_49 += kr_49.predict(X_test_49) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_kr_49, target)))
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_ridge_49 = np.zeros(train_shape)
predictions_ridge_49 = np.zeros(len(X_test_49))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_49[trn_idx]
tr_y = y_train[trn_idx]
ridge_49 = Ridge(alpha=6)
ridge_49.fit(tr_x,tr_y)
oof_ridge_49[val_idx] = ridge_49.predict(X_train_49[val_idx])
predictions_ridge_49 += ridge_49.predict(X_test_49) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_ridge_49, target)))
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_br_49 = np.zeros(train_shape)
predictions_br_49 = np.zeros(len(X_test_49))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_49[trn_idx]
tr_y = y_train[trn_idx]
br_49 = br()
br_49.fit(tr_x,tr_y)
oof_br_49[val_idx] = br_49.predict(X_train_49[val_idx])
predictions_br_49 += br_49.predict(X_test_49) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_br_49, target)))
folds = KFold(n_splits=5, shuffle=True, random_state=13)
oof_en_49 = np.zeros(train_shape)
predictions_en_49 = np.zeros(len(X_test_49))
#
for fold_, (trn_idx, val_idx) in enumerate(folds.split(X_train_49, y_train)):
print("fold n°{}".format(fold_+1))
tr_x = X_train_49[trn_idx]
tr_y = y_train[trn_idx]
en_49 = en(alpha=1.0,l1_ratio=0.05)
en_49.fit(tr_x,tr_y)
oof_en_49[val_idx] = en_49.predict(X_train_49[val_idx])
predictions_en_49 += en_49.predict(X_test_49) / folds.n_splits
print("CV score: {:<8.8f}".format(mean_squared_error(oof_en_49, target)))
再進行融合:
train_stack4 = np.vstack([oof_br_49,oof_kr_49,oof_en_49,oof_ridge_49]).transpose()
test_stack4 = np.vstack([predictions_br_49, predictions_kr_49,predictions_en_49,predictions_ridge_49]).transpose()
folds_stack = RepeatedKFold(n_splits=5, n_repeats=2, random_state=7)
oof_stack4 = np.zeros(train_stack4.shape[0])
predictions_lr4 = np.zeros(test_stack4.shape[0])
for fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack4,target)):
print("fold {}".format(fold_))
trn_data, trn_y = train_stack4[trn_idx], target.iloc[trn_idx].values
val_data, val_y = train_stack4[val_idx], target.iloc[val_idx].values
#LinearRegression
lr4 = lr()
lr4.fit(trn_data, trn_y)
oof_stack4[val_idx] = lr4.predict(val_data)
predictions_lr4 += lr4.predict(test_stack1) / 10
mean_squared_error(target.values, oof_stack4)
0.49491439094008133
模型融合
我們對263、383、49特徵的資料總共構建了4個模型,當前就需要對這些模型的預測結果進行融合,也可以通過學習一個迴歸來進行融合:
train_stack5 = np.vstack([oof_stack1,oof_stack2,oof_stack3,oof_stack4]).transpose()
test_stack5 = np.vstack([predictions_lr1, predictions_lr2,predictions_lr3,predictions_lr4]).transpose()
folds_stack = RepeatedKFold(n_splits=5, n_repeats=2, random_state=7)
oof_stack5 = np.zeros(train_stack5.shape[0])
predictions_lr5= np.zeros(test_stack5.shape[0])
for fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack5,target)):
print("fold {}".format(fold_))
trn_data, trn_y = train_stack5[trn_idx], target.iloc[trn_idx].values
val_data, val_y = train_stack5[val_idx], target.iloc[val_idx].values
#LinearRegression
lr5 = lr()
lr5.fit(trn_data, trn_y)
oof_stack5[val_idx] = lr5.predict(val_data)
predictions_lr5 += lr5.predict(test_stack5) / 10
mean_squared_error(target.values, oof_stack5)
0.4480223491250565
可以看到結果改進了不少。
結果儲存
submit_example = pd.read_csv('submit_example.csv',sep=',',encoding='latin-1')
submit_example['happiness'] = predictions_lr5
submit_example.loc[submit_example['happiness']>4.96,'happiness']= 5
submit_example.loc[submit_example['happiness']<=1.04,'happiness']= 1
submit_example.loc[(submit_example['happiness']>1.96)&(submit_example['happiness']<2.04),'happiness']= 2
# 進行整數解的近似
submit_example.to_csv("submision.csv",index=False)
整合學習案例2——蒸汽量預測
背景介紹
鍋爐的燃燒效率的影響因素很多,包括鍋爐的可調引數,如燃燒給量,一二次風,引風,返料風,給水水量;以及鍋爐的工況,比如鍋爐床溫、床壓,爐膛溫度、壓力,過熱器的溫度等。我們如何使用以上的資訊,根據鍋爐的工況,預測產生的蒸汽量呢?
資料分成訓練資料(train.txt)和測試資料(test.txt),其中欄位」V0」-「V37」,這38個欄位是作為特徵變數,」target」作為目標變數。我們需要利用訓練資料訓練出模型,預測測試資料的目標變數。
最終的評價指標為均方誤差MSE
具體程式碼
匯入需要的包
import warnings
warnings.filterwarnings("ignore") # 忽略各種不影響正常執行的警告
import matplotlib.pyplot as plt
import seaborn as sns
# 模型
import pandas as pd
import numpy as np
from scipy import stats
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV, RepeatedKFold, cross_val_score,cross_val_predict,KFold
from sklearn.metrics import make_scorer,mean_squared_error
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet
from sklearn.svm import LinearSVR, SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor,AdaBoostRegressor
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures,MinMaxScaler,StandardScaler
載入資料
data_train = pd.read_csv("train.txt", sep='\t')
data_test = pd.read_csv("test.txt", sep = '\t')
# 對訓練和測試的資料集疊在一起,對特徵處理的時候比較方便
data_train['oringin'] = 'train'
data_test['oringin'] = 'test'
data_all = pd.concat([data_train, data_test], axis = 0, ignore_index = True)
# 按照上下的方式疊在一起,並且忽略掉原來的索引,疊後重新建立索引
data_all.head()
data_all.tail() # target列自動填充NaN
探索資料分佈
這裡因為是感測器的資料,即連續變數,所以使用 kdeplot(核密度估計圖) 進行資料的初步分析,即EDA:
for column in data_all.columns[0:-2]:
# 為每個特徵畫一個核密度估計圖
g = sns.kdeplot(data_all[column][(data_all['oringin'] == "train")], color='Red',
shade=True) # 在曲線下面繪製陰影
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "test")], color='Blue',
ax=g, shade=True) # ax=g還是在原來的畫柄上
g.set_xlabel(column)
g.set_ylabel("Frequency")
g = g.legend(["train", "test"])
plt.show()
這裡為每個特徵都畫出了一個核密度的圖,太多了我就放一個吧。
那麼我們觀察所有的圖片,可以發現某些特徵的訓練資料和測試資料的分佈存在很大的差異,例如下圖:
那我們的選擇是刪除這些資料:
# 從以上的圖中可以看出特徵"V5","V9","V11","V17","V22","V28"中
# 訓練集資料分佈和測試集資料分佈不均,所以我們刪除這些特徵資料
for column in ["V5","V9","V11","V17","V22","V28"]:
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "train")], color="Red", shade = True)
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "test")], ax =g, color="Blue", shade= True)
g.set_xlabel(column)
g.set_ylabel("Frequency")
g = g.legend(["train","test"])
plt.show()
data_all.drop(["V5","V9","V11","V17","V22","V28"],axis=1,inplace=True)
# inplace 代表在原來的資料上做刪除操作
剩下的特徵,我們可以檢視彼此之間的相關性:
# 檢視特徵之之間的相關性
data_train1 = data_all[data_all["oringin"] == "train"].drop("oringin", axis=1)
plt.figure(figsize=(25,20))
colnm = data_train1.columns.tolist() # 得到特徵名稱構成的列表
mcorr = data_train1[colnm].corr(method="spearman") # 計算特徵之間的相關係數矩陣
mask = np.zeros_like(mcorr, dtype=np.bool) # 構造與mcorr同維度的矩陣,bool型別
mask[np.triu_indices_from(mask)] = True # 裡面那個是返回方陣的上三角的索引
cmap = sns.diverging_palette(220, 10, as_cmap = True)
# 從220和10之間建立一個逐漸變化色板物件
g = sns.heatmap(mcorr, mask = mask, cmap = cmap, square = True,
annot = True, fmt='0.2f')
# 第一個為資料集,cmap為顏色條的名稱或者物件或者列表,annot代表是否寫入數值,
# square將每個單元格設定為方形
plt.show()
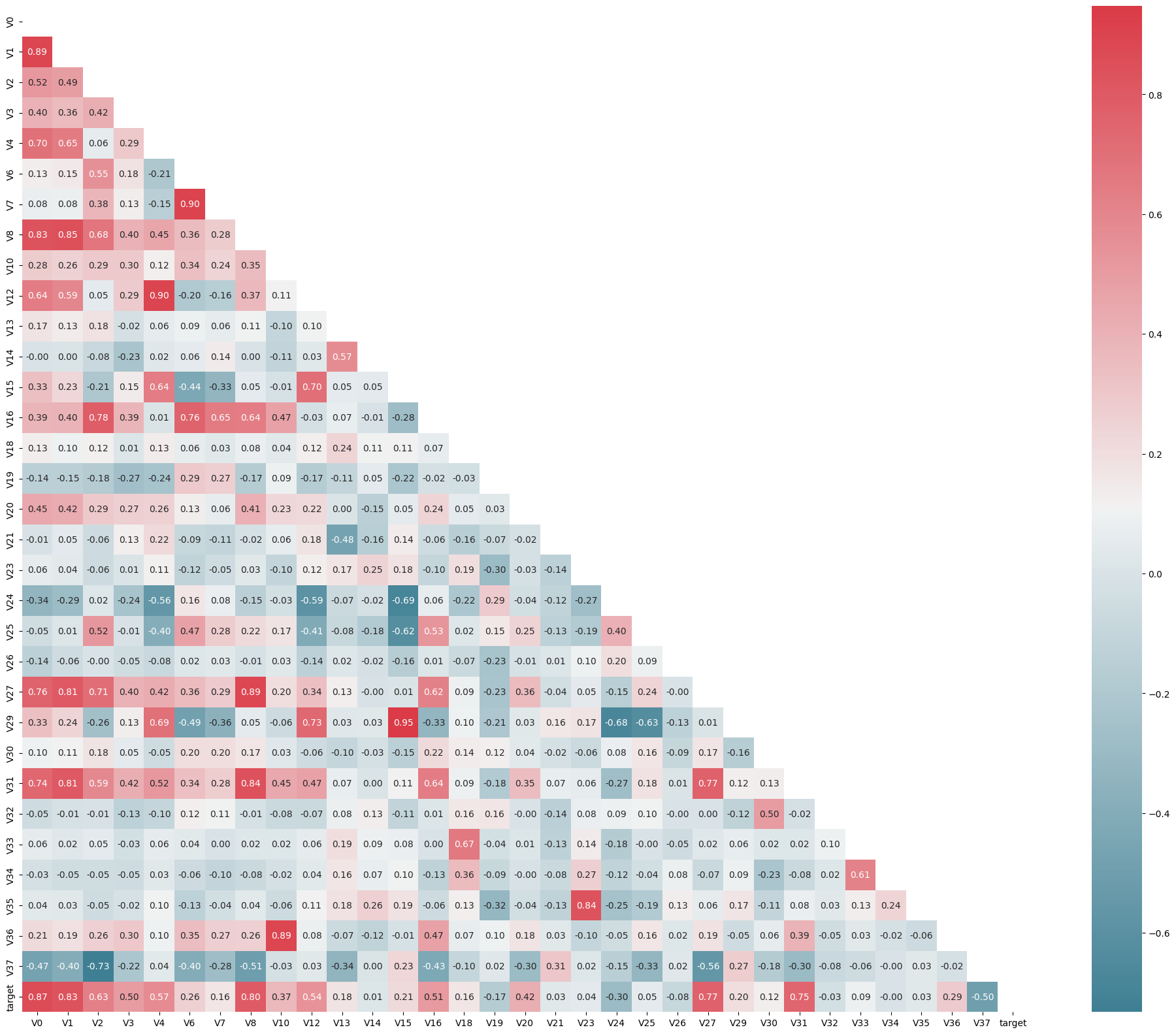
可以看到部分特徵之間的相關性較強。而部分特徵與標籤列的相關性太小,可以認為這些特徵的貢獻很有限,因此可以進行刪除:
# 將與target相關性過小的特徵進行刪除
threshold = 0.1
corr_matrix = data_train1.corr().abs() # 相關係數矩陣的絕對值
drop_col = corr_matrix[corr_matrix["target"] < threshold ].index
print(drop_col)
data_all.drop(drop_col, axis=1, inplace=True)
Index(['V14', 'V21', 'V25', 'V26', 'V32', 'V33', 'V34'], dtype='object')
接下來需要對資料進行歸一化操作:
# 進行歸一化操作
cols_numeric = list(data_all.columns) # 當前的特徵列表
cols_numeric.remove("oringin") # 拿掉這一列
def scale_minmax(col):
return (col-col.min()) / (col.max() - col.min())
scale_cols = [col for col in cols_numeric if col != "target"] # 除了目標,相當於remove
data_all[scale_cols] = data_all[scale_cols].apply(scale_minmax, axis = 0)
data_all[scale_cols].describe()
特徵工程
這裡引入了一個Box-Cox變換,因為大部分假設對資料有一定的要求,那我們希望資料的特徵能夠儘量滿足正態分佈的關係,而ox-cox變換的目標有兩個:一個是變換後,可以一定程度上減小不可觀測的誤差和預測變數的相關性。主要操作是對因變數轉換,使得變換後的因變數於迴歸自變數具有線性相依關係,誤差也服從正態分佈,誤差各分量是等方差且相互獨立。第二個是用這個變換來使得因變數獲得一些性質,比如在時間序列分析中的平穩性,或者使得因變數分佈為正態分佈。
下面我們可以先觀察變換前和變換後的區別:
fcols = 6
frows = len(cols_numeric) - 1 # 因為target不用
plt.figure(figsize = (4*fcols, 4*frows))
i = 0
for var in cols_numeric:
if var != "target":
data_tem = data_all[[var, "target"]].dropna() # 取出當前列和taget列並捨棄缺失值
# 畫處理前分佈圖
i+=1
plt.subplot(frows, fcols, i)
sns.distplot(data_tem[var], fit = stats.norm)
# 畫出直方圖,並且用stats.norm來進行擬合
plt.title(var+"oringinal")
plt.xlabel("")
# 畫QQ圖,看資料分佈是否服從正態分佈
i+=1
plt.subplot(frows, fcols, i)
_ = stats.probplot(data_tem[var],plot = plt)
plt.title("skew=" + "{:.4f}".format(stats.skew(data_tem[var])))
# stats.skew是計算偏度
plt.xlabel("")
plt.ylabel("")
# 畫散點圖
i+=1
plt.subplot(frows, fcols, i)
plt.plot(data_tem[var], data_tem["target"], ".", alpha = 0.5)
# 橫軸為var特徵,縱軸為target,用.來畫,透明度為0.5
plt.title("corr=" + "{:.2f}".format(np.corrcoef(data_tem[var],
data_tem["target"])[0][1]))
# 畫boxcox變換後的直方圖
i+=1
plt.subplot(frows, fcols, i)
trans_var, lambda_var = stats.boxcox(data_tem[var].dropna() + 1)
trans_var = scale_minmax(trans_var)
sns.distplot(trans_var, fit = stats.norm)
plt.title(var+"Transformed")
plt.xlabel("")
# 畫處理後的QQ圖
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(trans_var, plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(trans_var)))
plt.xlabel('')
plt.ylabel('')
# 畫處理後的散點圖
i+=1
plt.subplot(frows,fcols,i)
plt.plot(trans_var, data_tem['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(trans_var,
data_tem['target'])[0][1]))
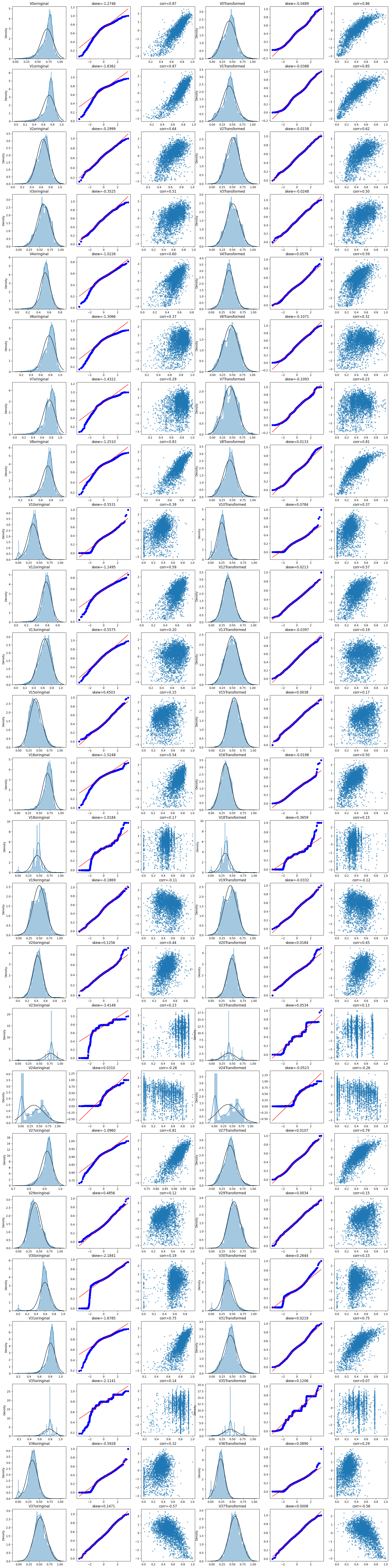
從QQ圖中可以看到經過變換後,其正態性好了很多!
因此對總體進行變換:
# 進行boxcox變換
cols_transform = data_all.columns[0:-2] # 這些是需要變換的特徵
for col in cols_transform:
data_all.loc[:, col], _ = stats.boxcox(data_all.loc[:,col] + 1)
# 因為我們已經標準化到-1與+1之間,而該函數要求輸入資料是正的,因此+1
print(data_all.target.describe())
count 2888.000000
mean 0.126353
std 0.983966
min -3.044000
25% -0.350250
50% 0.313000
75% 0.793250
max 2.538000
Name: target, dtype: float64
而對於標籤列來說也是如此,我們同樣可以對其嘗試進行變換,使用對數變換target目標值提升特徵資料的正太性:
# 變化之前
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_all.target.dropna(), fit = stats.norm)
plt.subplot(1,2,2)
_ = stats.probplot(data_all.target.dropna(), plot = plt)
# 掉target使用對數變化來提升其正態性質
sp = data_train.target
data_train.target1 = np.power(1.5, sp)
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_train.target1.dropna(),fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_train.target1.dropna(), plot=plt)
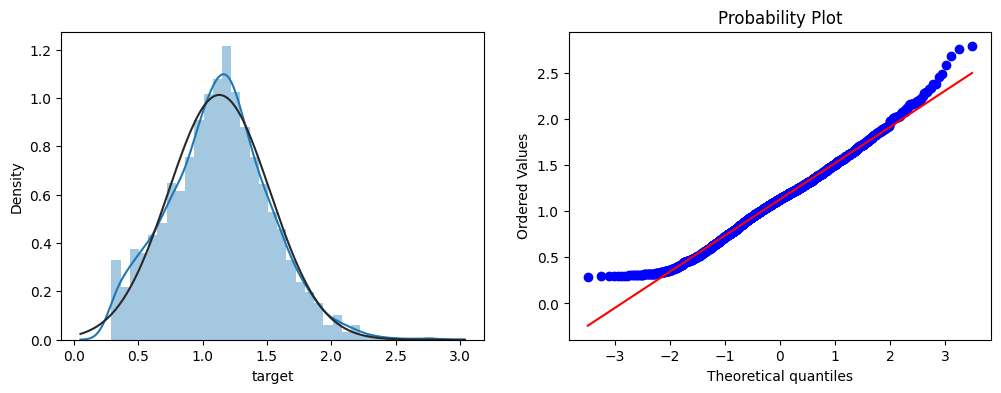
模型構建與整合學習
構建資料集
# 構建訓練集與測試集
from sklearn.model_selection import train_test_split
def get_training_data():
df_train = data_all[data_all["oringin"] == "train"]
df_train["label"] = data_train.target1 # 這是經過對數變化的標籤
y = df_train.target
X = df_train.drop(["oringin","target","label"],axis =1)
X_train, X_valid, y_train, y_valid = train_test_split(X, y, test_size = 0.3,
random_state = 100)
return X_train, X_valid, y_train, y_valid
def get_test_data():
df_test = data_all[data_all["oringin"] == "test"].reset_index(drop=True)
# 因為獲取的測試集的標籤是後面的,那麼需要從零開始,因此重置
return df_test.drop(["oringin", "target"], axis = 1)
構建評估函數
# 自己實現評估函數
from sklearn.metrics import make_scorer
def rmse(y_true, y_pred):
diff = y_pred - y_true
sum_sq = sum(diff * 2)
n = len(y_pred)
return np.sqrt(sum_sq / n)
def mse(y_true, y_pred):
return mean_squared_error(y_true, y_pred)
rmse_scorer = make_scorer(rmse, greater_is_better = False)
# 構建損失函數物件,第二個引數為True代表值越大越好,False代表值越小越好
mse_scorer = make_scorer(mse, greater_is_better = False)
構建尋找離群值的函數
# 該函數用來尋找離群值
def find_outliers(model, X, y, sigma = 3):
model.fit(X, y)
y_pred = pd.Series(model.predict(X), index = y.index)
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
z = (resid - mean_resid) / std_resid # 將差距標準化
outliers = z[abs(z) > sigma].index # 那些離平均差距超過sigma的
print("R2 = ", model.score(X, y))
print("rmse = ",rmse(y, y_pred))
print("mse = ", mean_squared_error(y, y_pred))
print("--" * 20)
print("mean of residuals:", mean_resid)
print("std of residuals:", std_resid)
print("--" * 20)
print(len(outliers), "outliers:")
print(outliers.tolist())
plt.figure(figsize = (15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y, y_pred,'.')
plt.plot(y.loc[outliers], y_pred.loc[outliers], "ro")
plt.legend(["Accepted","Outlier"])
plt.xlabel("y")
plt.ylabel("y_pred")
ax_132 = plt.subplot(1,3,2)
plt.plot(y, y- y_pred, ".")
plt.plot(y.loc[outliers], y.loc[outliers] - y_pred.loc[outliers], "ro")
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y - y_pred');
ax_133 = plt.subplot(1,3,3)
z.plot.hist(bins =50, ax = ax_133) # 畫出z的直方圖
z.loc[outliers].plot.hist(color='r', bins = 50, ax = ax_133)
plt.legend(['Accepted','Outlier'])
plt.xlabel('z')
return outliers
嘗試用嶺迴歸去尋找離散值:
X_train, X_valid, y_train, y_valid = get_training_data()
test = get_test_data()
outliers = find_outliers(Ridge(), X_train, y_train)
# 先用簡單的嶺迴歸去擬合試試看
X_outliers=X_train.loc[outliers]
y_outliers=y_train.loc[outliers]
X_t=X_train.drop(outliers)
y_t=y_train.drop(outliers)
R2 = 0.8766692297367809
rmse = nan
mse = 0.12180705697820839
----------------------------------------
mean of residuals: 1.8875439448847292e-16
std of residuals: 0.3490950551088699
----------------------------------------
22 outliers:
[2655, 2159, 1164, 2863, 1145, 2697, 2528, 2645, 691, 1085, 1874, 2647, 884, 2696, 2668, 1310, 1901, 1458, 2769, 2002, 2669, 1972]
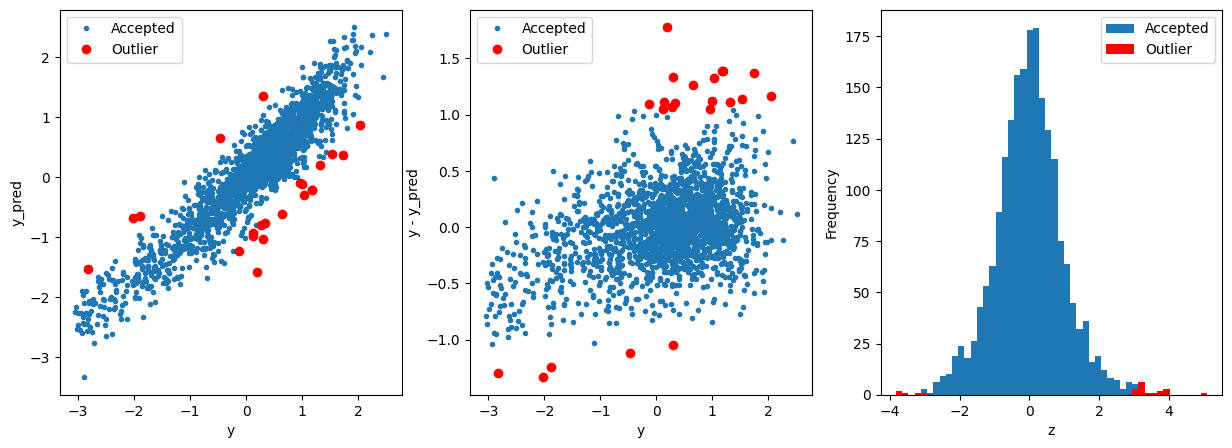
可以看到效果很好。
模型訓練
def get_trainning_data_omitoutliers():
#獲取訓練資料省略異常值
y=y_t.copy()
X=X_t.copy()
return X,y
def train_model(model, param_grid = [], X = [], y = [], splits = 5, repeats = 5):
# 嘗試實現訓練函數
if(len(y) == 0):
X, y = get_trainning_data_omitoutliers()
# 交叉驗證
rkfold = RepeatedKFold(n_splits = splits, n_repeats = repeats)
# 網格搜尋引數
if(len(param_grid) > 0):
gsearch = GridSearchCV(model, param_grid, cv = rkfold,
scoring = "neg_mean_squared_error",
verbose = 1, return_train_score = True)
gsearch.fit(X, y)
model = gsearch.best_estimator_
best_idx = gsearch.best_index_ # 最佳引數在param_gird中的索引
# 獲取交叉驗證評價指標
grid_results = pd.DataFrame(gsearch.cv_results_)
cv_mean = abs(grid_results.loc[best_idx, "mean_test_score"])
cv_std = grid_results.loc[best_idx, "std_test_score"]
else: # 不進行網格搜尋
grid_results = []
cv_results = cross_val_score(model, X ,y, scoring = "neg_mean_squared_error",
cv =rkfold)
cv_mean = abs(np.mean(cv_results))
cv_std = np.std(cv_results)
# 合併資料
cv_score = pd.Series({"mean":cv_mean, "std":cv_std})
# 預測
y_pred = model.predict(X)
# 模型效能的統計資料
print('----------------------')
print(model)
print('----------------------')
print('score=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print('mse=',mse(y, y_pred))
print('cross_val: mean=',cv_mean,', std=',cv_std)
# 殘差分析與視覺化
y_pred = pd.Series(y_pred,index=y.index)
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
z = (resid - mean_resid)/std_resid
n_outliers = sum(abs(z)>3)
outliers = z[abs(z)>3].index
return model, cv_score, grid_results
用嶺迴歸來進行擬合:
# 定義訓練變數儲存資料
opt_models = dict()
score_models = pd.DataFrame(columns=['mean','std'])
splits=5
repeats=5
model = 'RandomForest' #可替換,見案例分析一的各種模型
opt_models[model] = Ridge() #可替換,見案例分析一的各種模型
alph_range = np.arange(0.25,6,0.25)
param_grid = {'alpha': alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model],
param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),
abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')
Ridge(alpha=0.25)
----------------------
score= 0.8926884445023296
rmse= 5.109572960503552e-08
mse= 0.10540676395670681
cross_val: mean= 0.10910070303768438 , std= 0.005867873719733897
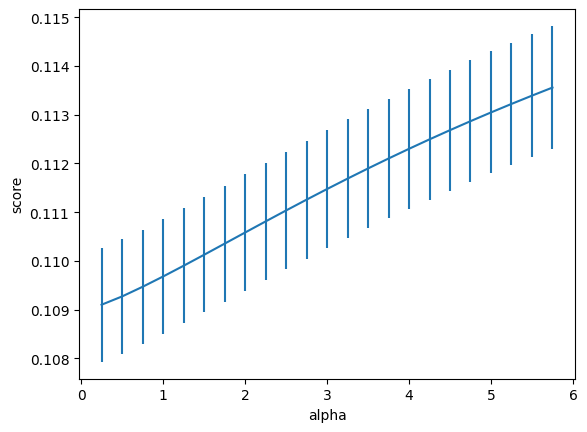
修改模型
我認為嶺迴歸比較簡單,因此嘗試將模型改為隨機森林迴歸,但是一直跑不動,因此自己寫了比較簡單的訓練過程。
先大概給個範圍進行隨機網格搜尋:
from sklearn.model_selection import RandomizedSearchCV
n_estimators_range=[int(x) for x in np.linspace(start=50,stop=500,num=50)]
max_depth_range=[5,10,15]
max_depth_range.append(None)
min_samples_split_range=[2,5,10]
min_samples_leaf_range=[4,8]
random_forest_seed = 1
random_forest_hp_range={'n_estimators':n_estimators_range,
'max_depth':max_depth_range,
'min_samples_split':min_samples_split_range,
'min_samples_leaf':min_samples_leaf_range
}
X, y = get_trainning_data_omitoutliers()
random_forest_model_test_base=RandomForestRegressor()
random_forest_model_test_random=RandomizedSearchCV(estimator=random_forest_model_test_base,
param_distributions=random_forest_hp_range,
n_iter=200,
n_jobs=-1,
cv=5,
verbose=1,
random_state=random_forest_seed
)
random_forest_model_test_random.fit(X,y)
best_hp_now=random_forest_model_test_random.best_params_
print(best_hp_now)
{'n_estimators': 343, 'min_samples_split': 2, 'min_samples_leaf': 4, 'max_depth': None}
再根據這個引數,縮小範圍進行網格搜尋:
n_estimators_range=[int(x) for x in np.linspace(start=320,stop=370,num=10)]
max_depth_range=[5,10,15]
max_depth_range.append(None)
min_samples_split_range=[1,2,3,4,5]
min_samples_leaf_range=[2,3,4,5,6]
random_forest_seed = 1
random_forest_hp_range_2={'n_estimators':n_estimators_range,
'max_depth':max_depth_range,
'min_samples_split':min_samples_split_range,
'min_samples_leaf':min_samples_leaf_range
}
random_forest_model_test_2_base=RandomForestRegressor()
random_forest_model_test_2_random = GridSearchCV(estimator=random_forest_model_test_2_base,
param_grid=random_forest_hp_range_2,
cv=5,
verbose=1,
n_jobs=-1)
random_forest_model_test_2_random.fit(X,y)
best_hp_now_2=random_forest_model_test_2_random.best_params_
print(best_hp_now_2)
{'max_depth': None, 'min_samples_leaf': 2, 'min_samples_split': 3, 'n_estimators': 364}
因此接下來做出預測並儲存:
random_forest_model_final=random_forest_model_test_2_random.best_estimator_
y_predict = pd.DataFrame(random_forest_model_final.predict(test))
y_predict.to_csv("predict.txt", header = None,index = False)
完結!